GuaB3, an overlooked enzyme in cyanobacteria's toolbox that sheds light on IMP dehydrogenase evolution.
Hernandez-Gomez, A., Irisarri, I., Fernandez-Justel, D., Pelaez, R., Jimenez, A., Revuelta, J.L., Balsera, M., Buey, R.M.(2023) Structure 31: 1526-1534.e4
- PubMed: 37875114
- DOI: https://doi.org/10.1016/j.str.2023.09.014
- Primary Citation of Related Structures:
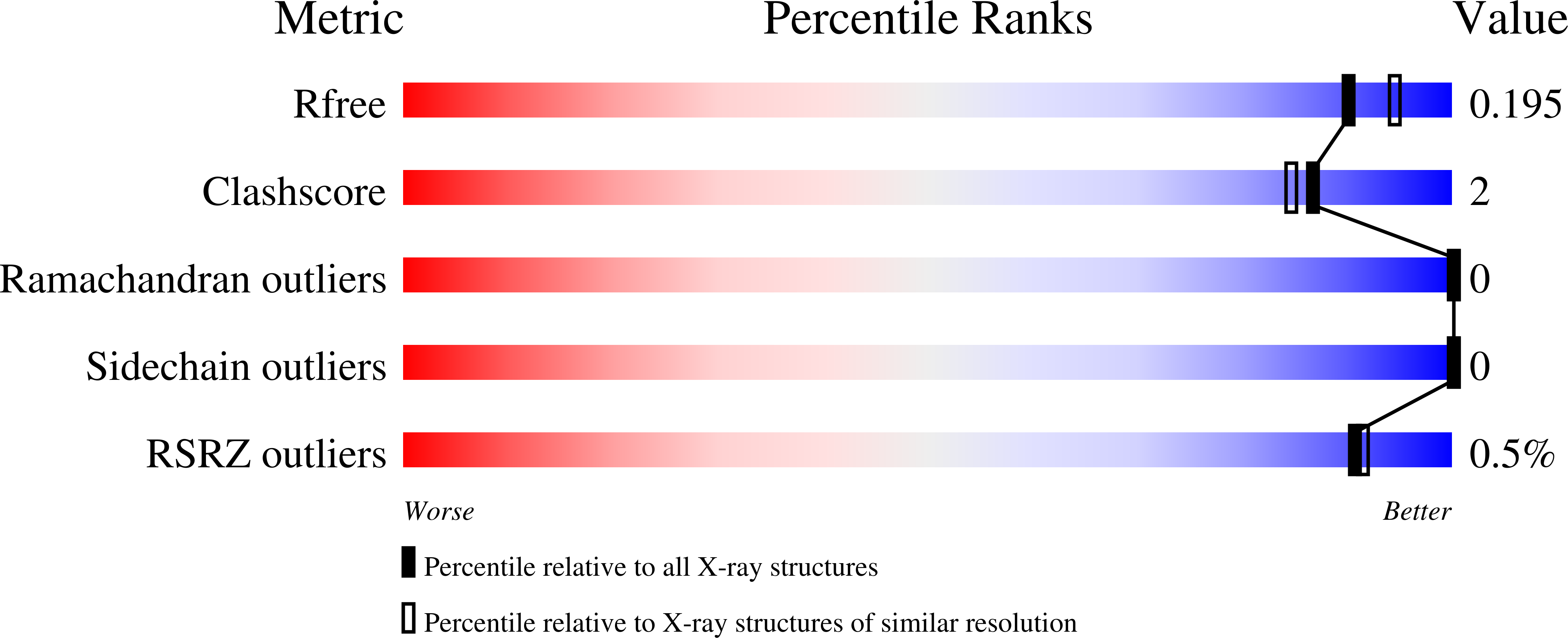
8P37, 8P4Q - PubMed Abstract:
IMP dehydrogenase and GMP reductase are enzymes from the same protein family with analogous structures and catalytic mechanisms that have gained attention because of their essential roles in nucleotide metabolism and as potential drug targets. This study focusses on GuaB3, a less-explored enzyme within this family. Phylogenetic analysis uncovers GuaB3's independent evolution from other members of the family and it predominantly occurs in Cyanobacteria. Within this group, GuaB3 functions as a unique IMP dehydrogenase, while its counterpart in Actinobacteria has a yet unknown function. Synechocystis sp. PCC6803 GuaB3 structures demonstrate differences in the active site compared to canonical IMP dehydrogenases, despite shared catalytic mechanisms. These findings highlight the essential role of GuaB3 in Cyanobacteria, provide insights into the diversity and evolution of the IMP dehydrogenase protein family, and reveal a distinctive characteristic in nucleotide metabolism, potentially aiding in combating harmful cyanobacterial blooms-a growing concern for humans and wildlife.
Organizational Affiliation:
Metabolic Engineering Group, Dpto. Microbiología y Genética, Universidad de Salamanca, Edificio Departamental, Campus Miguel de Unamuno, 37007 Salamanca, Spain.