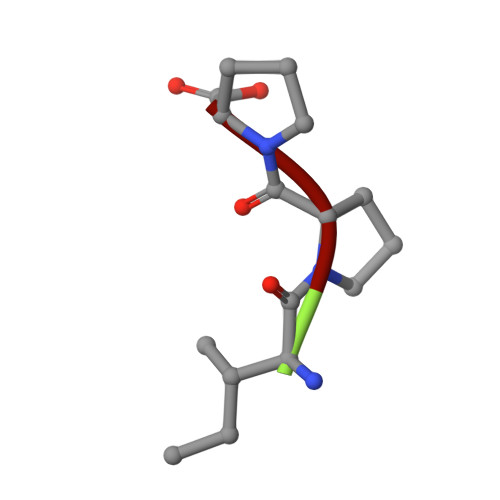
Structural insights into the inhibitory mechanism of angiotensin-I-converting enzyme by the lactotripeptides IPP and VPP.
Gregory, K.S., Cozier, G.E., Schwager, S.L.U., Sturrock, E.D., Acharya, K.R.(2024) FEBS Lett 598: 242-251
- PubMed: 37904282
- DOI: https://doi.org/10.1002/1873-3468.14768
- Primary Citation of Related Structures:
8QFX, 8QHL - PubMed Abstract:
Human somatic angiotensin-1-converting enzyme (sACE) is composed of a catalytic N-(nACE) and C-domain (cACE) of similar size with different substrate specificities. It is involved in the regulation of blood pressure by converting angiotensin I to the vasoconstrictor angiotensin II and has been a major focus in the development of therapeutics for hypertension. Bioactive peptides from various sources, including milk, have been identified as natural ACE inhibitors. We report the structural basis for the role of two lacototripeptides, Val-Pro-Pro and Ile-Pro-Pro, in domain-specific inhibition of ACE using X-ray crystallography and kinetic analysis. The lactotripeptides have preference for nACE due to altered polar interactions distal to the catalytic zinc ion. Elucidating the mechanism of binding and domain selectivity of these peptides also provides important insights into the functional roles of ACE.
Organizational Affiliation:
Department of Life Sciences, University of Bath, UK.