New Super-Slow Substrates as novel Sirtuin-Inhibitors
Friedrich, F., Kalbas, D., Einsle, O., Jung, M., Schutkowski, M.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| NAD-dependent protein deacetylase sirtuin-2 | 304 | Homo sapiens | Mutation(s): 0 Gene Names: SIRT2, SIR2L, SIR2L2 EC: 3.5.1 (PDB Primary Data), 2.3.1 (UniProt), 2.3.1.286 (UniProt) | 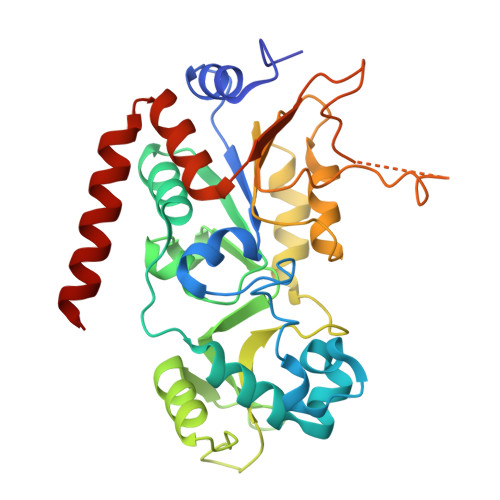 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q8IXJ6 (Homo sapiens) Explore Q8IXJ6 Go to UniProtKB: Q8IXJ6 | |||||
PHAROS: Q8IXJ6 GTEx: ENSG00000068903 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8IXJ6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Peptide-based super-slow substrate TNFn-6 | 10 | Homo sapiens | Mutation(s): 0 |  | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 8 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAD (Subject of Investigation/LOI) Query on NAD | K [auth A] | NICOTINAMIDE-ADENINE-DINUCLEOTIDE C21 H27 N7 O14 P2 BAWFJGJZGIEFAR-NNYOXOHSSA-N |  | ||
| WU8 (Subject of Investigation/LOI) Query on WU8 | M [auth B] | 3-dodecylsulfanyl-2,2-dimethyl-propanoic acid C17 H34 O2 S CBYMTUKZNNXAOB-UHFFFAOYSA-N |  | ||
| BTB Query on BTB | F [auth A] | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL C8 H19 N O5 OWMVSZAMULFTJU-UHFFFAOYSA-N |  | ||
| PGE Query on PGE | J [auth A] | TRIETHYLENE GLYCOL C6 H14 O4 ZIBGPFATKBEMQZ-UHFFFAOYSA-N |  | ||
| BU3 Query on BU3 | H [auth A] | (R,R)-2,3-BUTANEDIOL C4 H10 O2 OWBTYPJTUOEWEK-QWWZWVQMSA-N |  | ||
| DMS Query on DMS | G [auth A] | DIMETHYL SULFOXIDE C2 H6 O S IAZDPXIOMUYVGZ-UHFFFAOYSA-N |  | ||
| ZN Query on ZN | C [auth A] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | D [auth A], E [auth A], I [auth A], L [auth A] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| NIY Query on NIY | B | L-PEPTIDE LINKING | C9 H10 N2 O5 |  | TYR |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 38.056 | α = 90 |
| b = 74.847 | β = 96.37 |
| c = 55.981 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| autoPROC | data reduction |
| Aimless | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| German Research Foundation (DFG) | Germany | SFB 992 |
| German Research Foundation (DFG) | Germany | 295/18-1 |