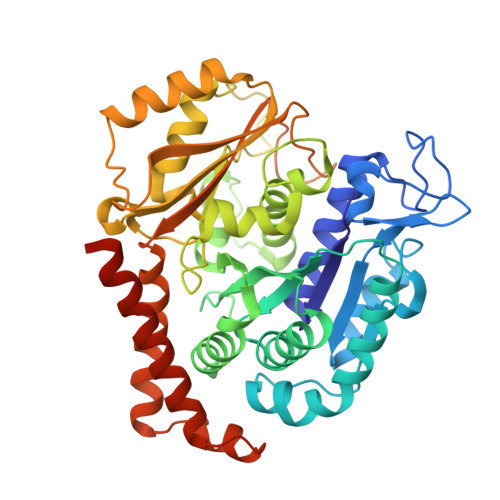
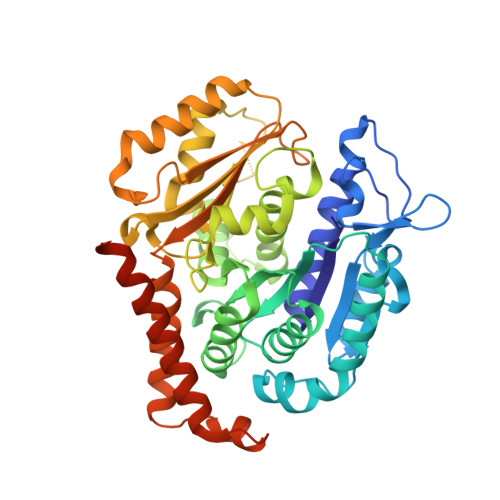
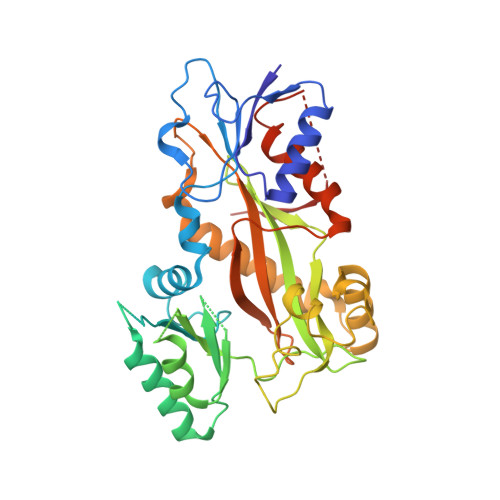
Bridging the maytansine and vinca sites: Cryptophycins target beta-tubulin's T5-loop.
Abel, A.C., Muhlethaler, T., Dessin, C., Schachtsiek, T., Sammet, B., Sharpe, T., Steinmetz, M.O., Sewald, N., Prota, A.E.(2024) J Biol Chem 300: 107363-107363
- PubMed: 38735475
- DOI: https://doi.org/10.1016/j.jbc.2024.107363
- Primary Citation of Related Structures:
8R67 - PubMed Abstract:
Cryptophycins are microtubule-targeting agents (MTAs) that belong to the most potent antimitotic compounds known to date; however, their exact molecular mechanism of action remains unclear. Here, we present the 2.2 Å resolution X-ray crystal structure of a potent cryptophycin derivative bound to the αβ-tubulin heterodimer. The structure addresses conformational issues present in a previous 3.3 Å resolution cryo-electron microscopy structure of cryptophycin-52 bound to the maytansine site of β-tubulin. It further provides atomic details on interactions of cryptophycins, which had not been described previously, including ones that are in line with structure-activity relationship (SAR) studies. Interestingly, we discovered a second cryptophycin-binding site that involves the T5-loop of β-tubulin, a critical secondary structure element involved in the exchange of the guanosine nucleotide and in the formation of longitudinal tubulin contacts in microtubules. Cryptophycins are the first natural ligands found to bind to this new "βT5-loop site" that bridges the maytansine and vinca sites. Our results offer unique avenues to rationally design novel MTAs with the capacity to modulate T5-loop dynamics and to simultaneously engage multiple β-tubulin binding sites.
Organizational Affiliation:
Laboratory of Biomolecular Research, Department of Biology and Chemistry, Paul Scherrer Institute, 5232 Villigen PSI, Switzerland; Biozentrum, University of Basel, Spitalstrasse 41, 4056 Basel, Switzerland.