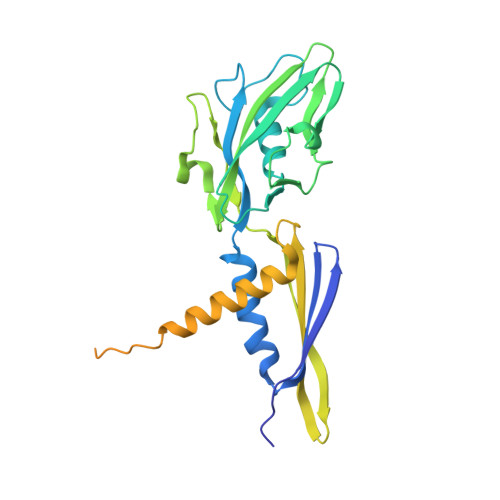
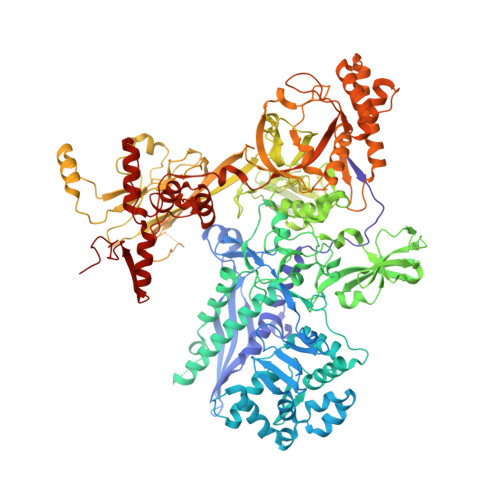
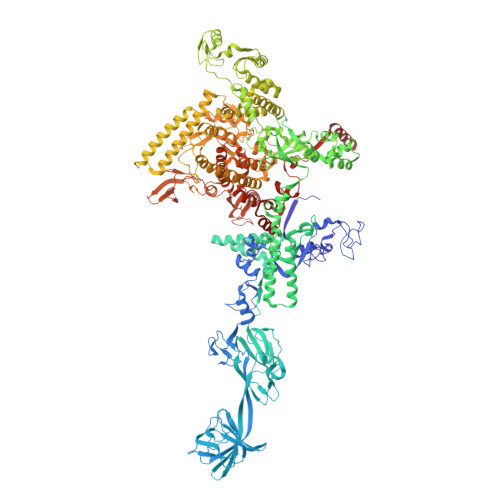
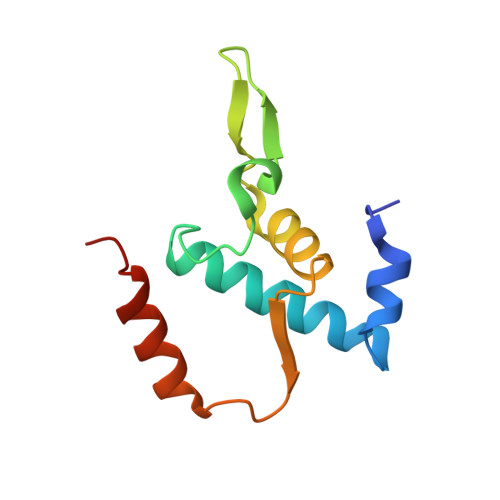
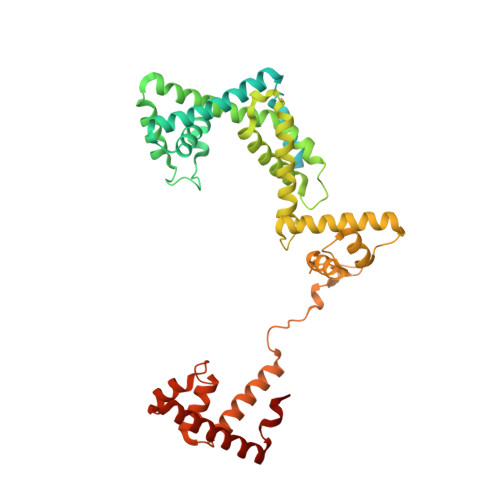
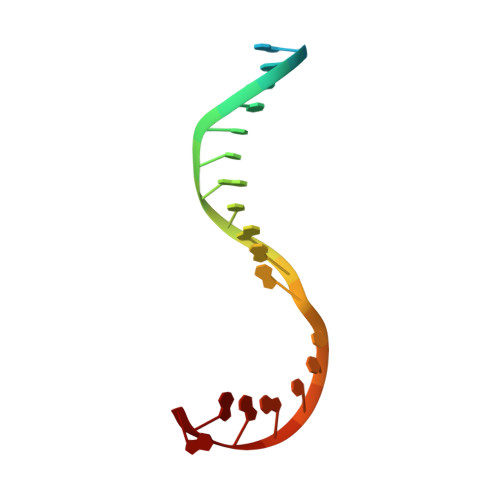
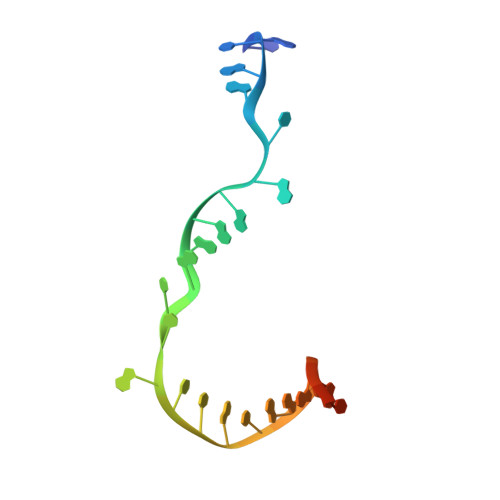
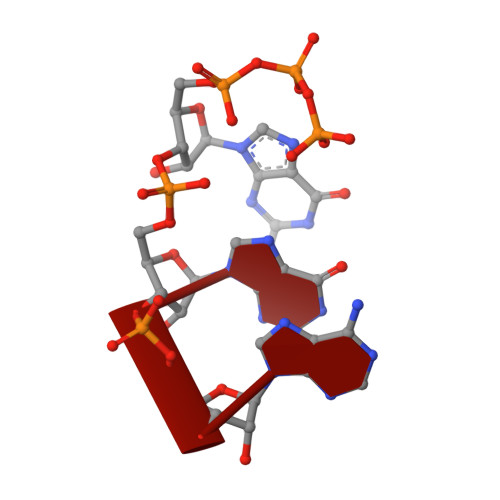
Nanopore tweezers show fractional-nucleotide translocation in sequence-dependent pausing by RNA polymerase.
Nova, I.C., Craig, J.M., Mazumder, A., Laszlo, A.H., Derrington, I.M., Noakes, M.T., Brinkerhoff, H., Yang, S., Vahedian-Movahed, H., Li, L., Zhang, Y., Bowman, J.L., Huang, J.R., Mount, J.W., Ebright, R.H., Gundlach, J.H.(2024) Proc Natl Acad Sci U S A 121: e2321017121-e2321017121
- PubMed: 38990947
- DOI: https://doi.org/10.1073/pnas.2321017121
- Primary Citation of Related Structures:
8W8N, 8W8O, 8W8P - PubMed Abstract:
RNA polymerases (RNAPs) carry out the first step in the central dogma of molecular biology by transcribing DNA into RNA. Despite their importance, much about how RNAPs work remains unclear, in part because the small (3.4 Angstrom) and fast (~40 ms/nt) steps during transcription were difficult to resolve. Here, we used high-resolution nanopore tweezers to observe the motion of single Escherichia coli RNAP molecules as it transcribes DNA ~1,000 times improved temporal resolution, resolving single-nucleotide and fractional-nucleotide steps of individual RNAPs at saturating nucleoside triphosphate concentrations. We analyzed RNAP during processive transcription elongation and sequence-dependent pausing at the yrbL elemental pause sequence. Each time RNAP encounters the yrbL elemental pause sequence, it rapidly interconverts between five translocational states, residing predominantly in a half-translocated state. The kinetics and force-dependence of this half-translocated state indicate it is a functional intermediate between pre- and post-translocated states. Using structural and kinetics data, we show that, in the half-translocated and post-translocated states, sequence-specific protein-DNA interaction occurs between RNAP and a guanine base at the downstream end of the transcription bubble (core recognition element). Kinetic data show that this interaction stabilizes the half-translocated and post-translocated states relative to the pre-translocated state. We develop a kinetic model for RNAP at the yrbL pause and discuss this in the context of key structural features.
Organizational Affiliation:
Department of Physics, University of Washington, Seattle, WA 98195.