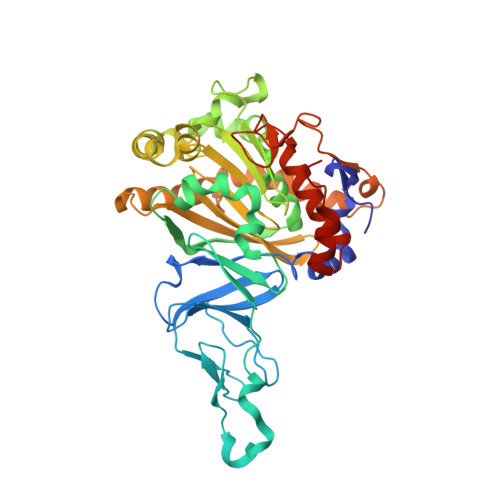
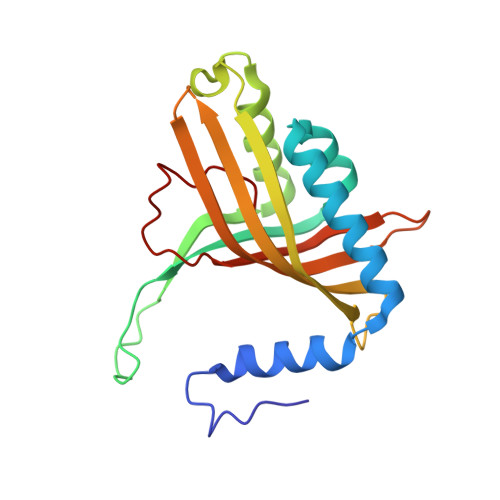
Structural insight into the dioxygenation of nitroarene compounds: the crystal structure of nitrobenzene dioxygenase.
Friemann, R., Ivkovic-Jensen, M.M., Lessner, D.J., Yu, C.L., Gibson, D.T., Parales, R.E., Eklund, H., Ramaswamy, S.(2005) J Mol Biol 348: 1139-1151
- PubMed: 15854650
- DOI: https://doi.org/10.1016/j.jmb.2005.03.052
- Primary Citation of Related Structures:
2BMO, 2BMQ, 2BMR - PubMed Abstract:
Nitroaromatic compounds are used extensively in many industrial processes and have been released into the environment where they are considered environmental pollutants. Nitroaromatic compounds, in general, are resistant to oxidative attack due to the electron-withdrawing nature of the nitro groups and the stability of the benzene ring. However, the bacterium Comamonas sp. strain JS765 can grow with nitrobenzene as a sole source of carbon, nitrogen and energy. Biodegradation is initiated by the nitrobenzene dioxygenase (NBDO) system. We have determined the structure of NBDO, which has a hetero-hexameric structure similar to that of several other Rieske non-heme iron dioxygenases. The catalytic subunit contains a Rieske iron-sulfur center and an active-site mononuclear iron atom. The structures of complexes with substrates nitrobenzene and 3-nitrotoluene reveal the structural basis for its activity with nitroarenes. The substrate pocket contains an asparagine residue that forms a hydrogen bond to the nitro-group of the substrate, and orients the substrate in relation to the active-site mononuclear iron atom, positioning the molecule for oxidation at the nitro-substituted carbon.
Organizational Affiliation:
Department of Molecular Biology, Swedish University of Agricultural Sciences, Uppsala Biomedical Center, Box 590 S-751 24 Uppsala, Sweden. [email protected]