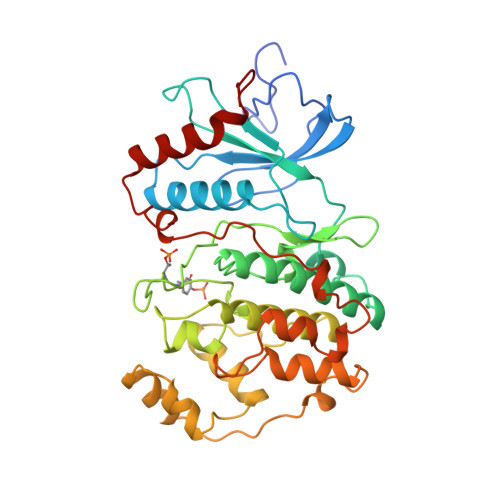
Activation mechanism of the MAP kinase ERK2 by dual phosphorylation.
Canagarajah, B.J., Khokhlatchev, A., Cobb, M.H., Goldsmith, E.J.(1997) Cell 90: 859-869
- PubMed: 9298898
- DOI: https://doi.org/10.1016/s0092-8674(00)80351-7
- Primary Citation of Related Structures:
2ERK - PubMed Abstract:
The structure of the active form of the MAP kinase ERK2 has been solved, phosphorylated on a threonine and a tyrosine residue within the phosphorylation lip. The lip is refolded, bringing the phosphothreonine and phosphotyrosine into alignment with surface arginine-rich binding sites. Conformational changes occur in the lip and neighboring structures, including the P+1 site, the MAP kinase insertion, the C-terminal extension, and helix C. Domain rotation and remodeling of the proline-directed P+1 specificity pocket account for the activation. The conformation of the P+1 pocket is similar to a second proline-directed kinase, CDK2-CyclinA, thus permitting the origin of this specificity to be defined. Conformational changes outside the lip provide loci at which the state of phosphorylation can be felt by other cellular components.
Organizational Affiliation:
Department of Biochemistry, The University of Texas Southwestern Medical Center at Dallas, 75235-9050, USA.