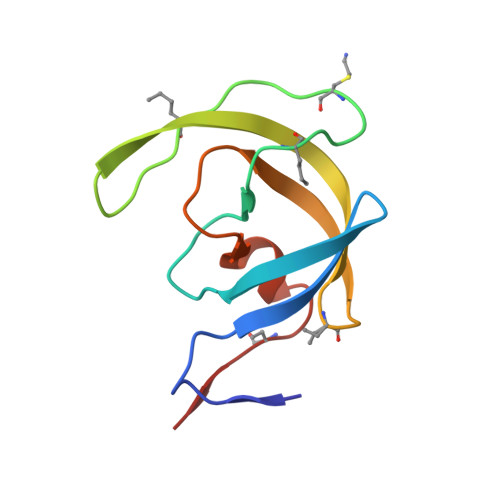
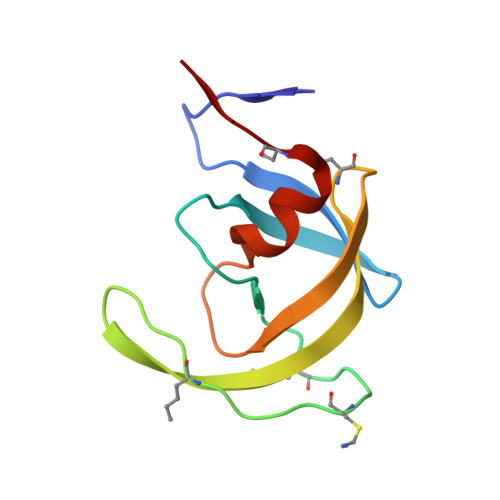
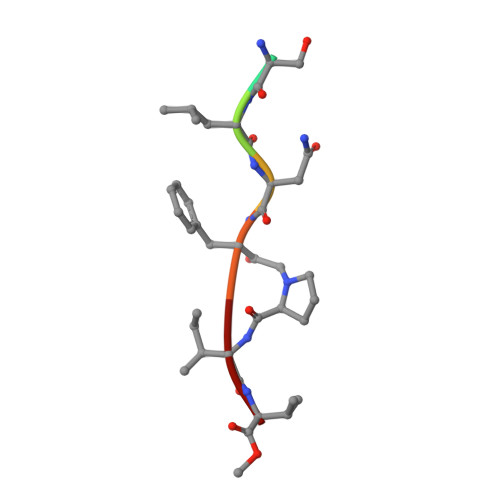
Insights from Atomic-Resolution X-Ray Structures of Chemically Synthesized HIV-1 Protease in Complex with Inhibitors.
Johnson, E.C.B., Malito, E., Shen, Y., Pentelute, B., Rich, D., Florian, J., Tang, W.J., Kent, S.B.H.(2007) J Mol Biol 373: 573
- PubMed: 17869270
- DOI: https://doi.org/10.1016/j.jmb.2007.07.054
- Primary Citation of Related Structures:
2J9J, 2J9K - PubMed Abstract:
The human immunodeficiency virus 1 (HIV-1) protease (PR) is an aspartyl protease essential for HIV-1 viral infectivity. HIV-1 PR has one catalytic site formed by the homodimeric enzyme. We chemically synthesized fully active HIV-1 PR using modern ligation methods. When complexed with the classic substrate-derived inhibitors JG-365 and MVT-101, the synthetic HIV-1 PR formed crystals that diffracted to 1.04- and 1.2-A resolution, respectively. These atomic-resolution structures revealed additional structural details of the HIV-1 PR's interactions with its active site ligands. Heptapeptide inhibitor JG-365, which has a hydroxyethylamine moiety in place of the scissile bond, binds in two equivalent antiparallel orientations within the catalytic groove, whereas the reduced isostere hexapeptide MVT-101 binds in a single orientation. When JG-365 was converted into the natural peptide substrate for molecular dynamic simulations, we found putative catalytically competent reactant states for both lytic water and direct nucleophilic attack mechanisms. Moreover, free energy perturbation calculations indicated that the insertion of catalytic water into the catalytic site is an energetically favorable process.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, The University of Chicago, IL 60637, USA.