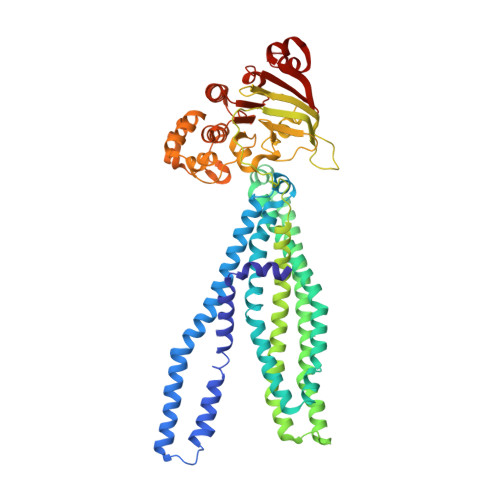
Structure of the multidrug ABC transporter Sav1866 from Staphylococcus aureus in complex with AMP-PNP.
Dawson, R.J.P., Locher, K.P.(2007) FEBS Lett 581: 935-938
- PubMed: 17303126
- DOI: https://doi.org/10.1016/j.febslet.2007.01.073
- Primary Citation of Related Structures:
2ONJ - PubMed Abstract:
Staphylococcus aureus Sav1866 is a bacterial homolog of the human ABC transporter Mdr1 that causes multidrug resistance in cancer cells. We report the crystal structure of Sav1866 in complex with adenosine-5'-(beta,gamma-imido)triphosphate (AMP-PNP) at 3.4A resolution and compare it with the previously determined structure of Sav1866 with bound ADP. Besides differences in the ATP-binding sites, no significant conformational changes were observed. The results confirm that the ATP-bound state of multidrug ABC transporters is coupled to an outward-facing conformation of the transmembrane domains.
Organizational Affiliation:
Institute of Molecular Biology and Biophysics, ETH Zurich HPK D14.3, 8093 Zurich, Switzerland.