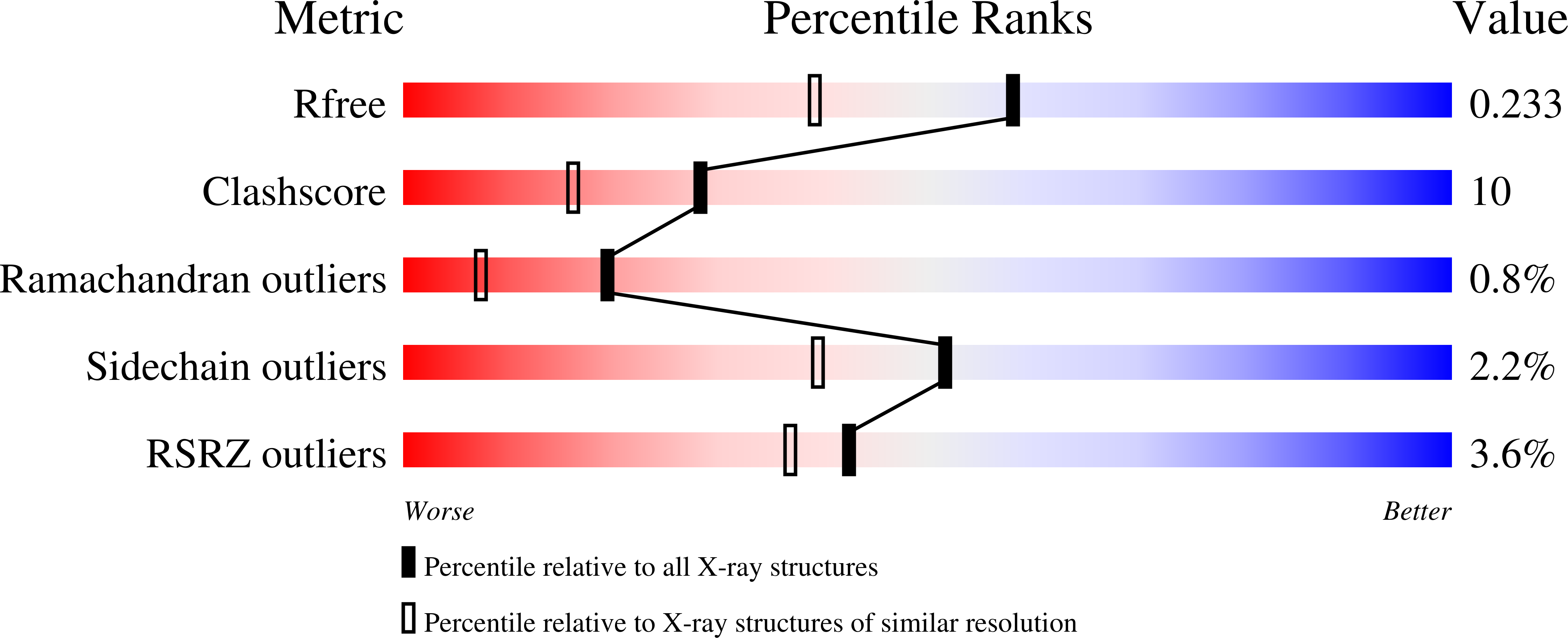
Crystal structure of hereditary vitamin D-resistant rickets--associated mutant H305Q of vitamin D nuclear receptor bound to its natural ligand
Rochel, N., Hourai, S., Moras, D.(2010) J Steroid Biochem Mol Biol 121: 84-87
- PubMed: 20403435
- DOI: https://doi.org/10.1016/j.jsbmb.2010.04.008
- Primary Citation of Related Structures:
3M7R - PubMed Abstract:
In the nuclear receptor of vitamin D (VDR) histidine 305 participates to the anchoring of the ligand. The VDR H305Q mutation was identified in a patient who exhibited the hereditary vitamin D-resistant rickets (HVDRR). We report the crystal structure of human VDR H305Q-ligand binding domain bound to 1alpha,25(OH)2D3 solved at 1.8A resolution. The protein adopts the active conformation of the wild-type liganded VDR. A local conformational flexibility at the mutation site weakens the hydrogen bond between the 25-OH with Gln305, thus explaining the lower affinity of the mutant proteins for calcitriol. The structure provides the basis for a rational approach to the design of more potent ligands for the treatment of HVDRR.
Organizational Affiliation:
IGBMC (Institut de Génétique et de Biologie Moléculaire et Cellulaire), Département de Biologie et de Génomique Structurales, Centre National de la Recherche Scientifique, Institut National de la Santé de la Recherche Médicale, Université de Strasbourg, 1 rue Laurent Fries, Illkirch, France. [email protected]