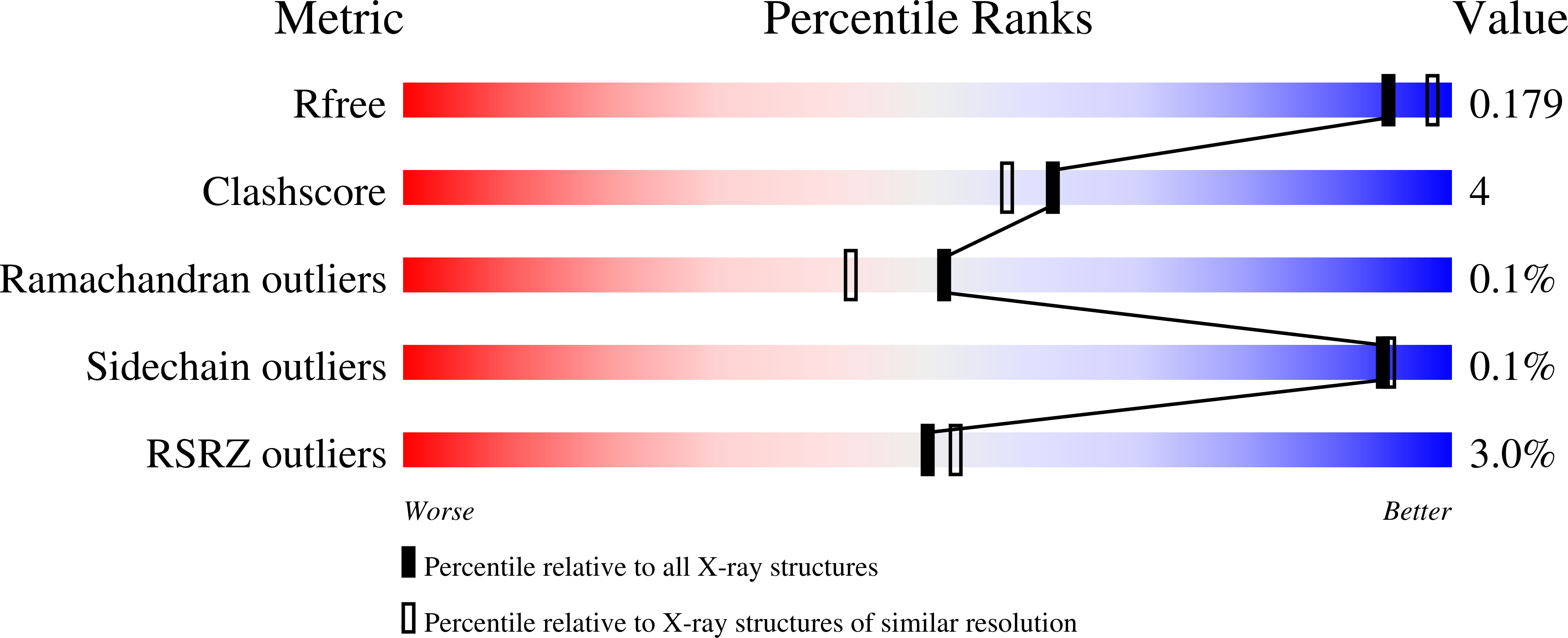
A novel inhibitor of Plasmodium falciparum spermidine synthase: a twist in the tail.
Burger, P.B., Williams, M., Sprenger, J., Reeksting, S.B., Botha, M., Muller, I.B., Joubert, F., Birkholtz, L.M., Louw, A.I.(2015) Malar J 14: 54-54
- PubMed: 25651815
- DOI: https://doi.org/10.1186/s12936-015-0572-z
- Primary Citation of Related Structures:
3RIE - PubMed Abstract:
Plasmodium falciparum is the most pathogenic of the human malaria parasite species and a major cause of death in Africa. It's resistance to most of the current drugs accentuates the pressing need for new chemotherapies. Polyamine metabolism of the parasite is distinct from the human pathway making it an attractive target for chemotherapeutic development. Plasmodium falciparum spermidine synthase (PfSpdS) catalyzes the synthesis of spermidine and spermine. It is a major polyamine flux-determining enzyme and spermidine is a prerequisite for the post-translational activation of P. falciparum eukaryotic translation initiation factor 5A (elF5A). The most potent inhibitors of eukaryotic SpdS's are not specific for PfSpdS. 'Dynamic' receptor-based pharmacophore models were generated from published crystal structures of SpdS with different ligands. This approach takes into account the inherent flexibility of the active site, which reduces the entropic penalties associated with ligand binding. Four dynamic pharmacophore models were developed and two inhibitors, (1R,4R)-(N1-(3-aminopropyl)-trans-cyclohexane-1,4-diamine (compound 8) and an analogue, N-(3-aminopropyl)-cyclohexylamine (compound 9), were identified. A crystal structure containing compound 8 was solved and confirmed the in silico prediction that its aminopropyl chain traverses the catalytic centre in the presence of the byproduct of catalysis, 5'-methylthioadenosine. The IC50 value of compound 9 is in the same range as that of the most potent inhibitors of PfSpdS, S-adenosyl-1,8-diamino-3-thio-octane (AdoDATO) and 4MCHA and 100-fold lower than that of compound 8. Compound 9 was originally identified as a mammalian spermine synthase inhibitor and does not inhibit mammalian SpdS. This implied that these two compounds bind in an orientation where their aminopropyl chains face the putrescine binding site in the presence of the substrate, decarboxylated S-adenosylmethionine. The higher binding affinity and lower receptor strain energy of compound 9 compared to compound 8 in the reversed orientation explained their different IC50 values. The specific inhibition of PfSpdS by compound 9 is enabled by its binding in the additional cavity normally occupied by spermidine when spermine is synthesized. This is the first time that a spermine synthase inhibitor is shown to inhibit PfSpdS, which provides new avenues to explore for the development of novel inhibitors of PfSpdS.
Organizational Affiliation:
Department of Biochemistry, UP Centre for Sustainable Malaria Control, Faculty of Natural and Agricultural Sciences, University of Pretoria, Private Bag X20, Hatfield, 0028, South Africa. [email protected].