Structural and mechanistic insights into guanylylation of RNA-splicing ligase RtcB joining RNA between 3'-terminal phosphate and 5'-OH.
Englert, M., Xia, S., Okada, C., Nakamura, A., Tanavde, V., Yao, M., Eom, S.H., Konigsberg, W.H., Soll, D., Wang, J.(2012) Proc Natl Acad Sci U S A 109: 15235-15240
- PubMed: 22949672
- DOI: https://doi.org/10.1073/pnas.1213795109
- Primary Citation of Related Structures:
4DWQ, 4DWR - PubMed Abstract:
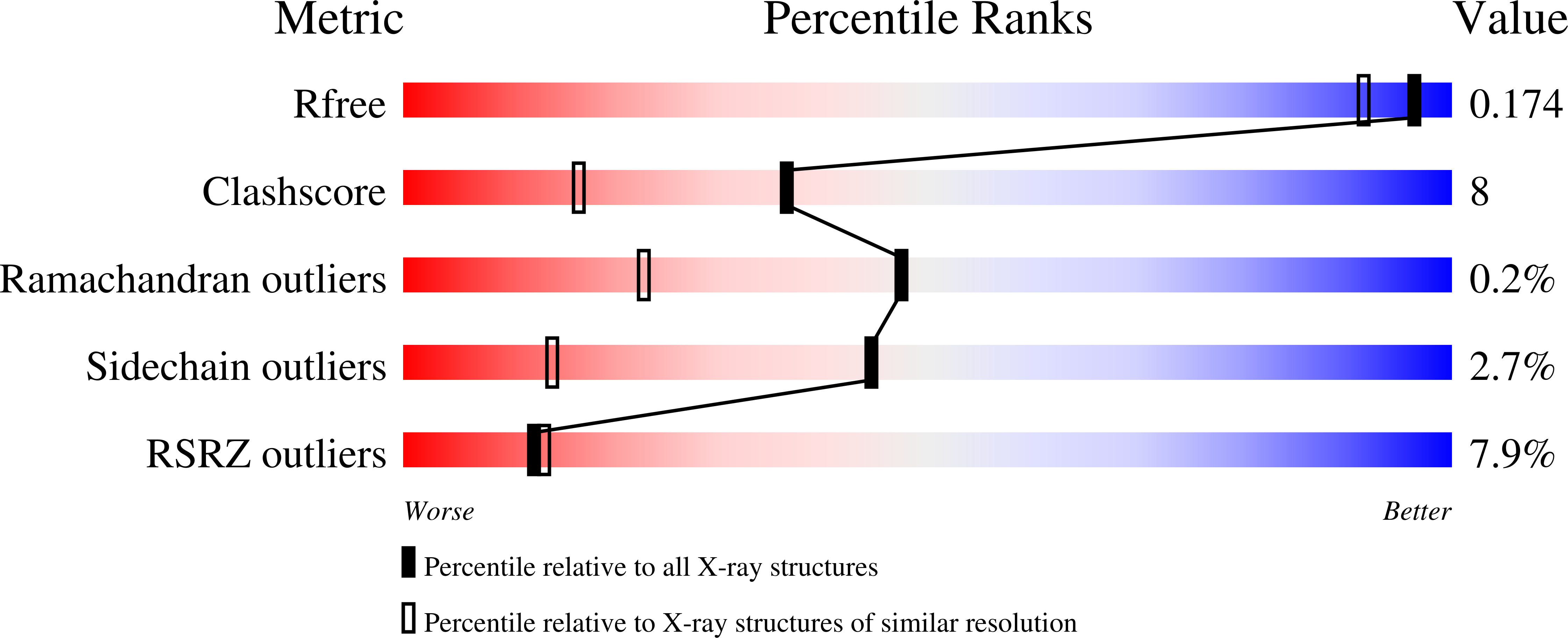
The RtcB protein has recently been identified as a 3'-phosphate RNA ligase that directly joins an RNA strand ending with a 2',3'-cyclic phosphate to the 5'-hydroxyl group of another RNA strand in a GTP/Mn(2+)-dependent reaction. Here, we report two crystal structures of Pyrococcus horikoshii RNA-splicing ligase RtcB in complex with Mn(2+) alone (RtcB/ Mn(2+)) and together with a covalently bound GMP (RtcB-GMP/Mn(2+)). The RtcB/ Mn(2+) structure (at 1.6 Å resolution) shows two Mn(2+) ions at the active site, and an array of sulfate ions nearby that indicate the binding sites of the RNA phosphate backbone. The structure of the RtcB-GMP/Mn(2+) complex (at 2.3 Å resolution) reveals the detailed geometry of guanylylation of histidine 404. The critical roles of the key residues involved in the binding of the two Mn(2+) ions, the four sulfates, and GMP are validated in extensive mutagenesis and biochemical experiments, which also provide a thorough characterization for the three steps of the RtcB ligation pathway: (i) guanylylation of the enzyme, (ii) guanylyl-transfer to the RNA substrate, and (iii) overall ligation. These results demonstrate that the enzyme's substrate-induced GTP binding site and the putative reactive RNA ends are in the vicinity of the binuclear Mn(2+) active center, which provides detailed insight into how the enzyme-bound GMP is tansferred to the 3'-phosphate of the RNA substrate for activation and subsequent nucleophilic attack by the 5'-hydroxyl of the second RNA substrate, resulting in the ligated product and release of GMP.
Organizational Affiliation:
Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT 06520, USA.