Structural insights into Gemin5-guided selection of pre-snRNAs for snRNP assembly
Xu, C., Ishikawa, H., Izumikawa, K., Li, L., He, H., Nobe, Y., Yamauchi, Y., Shahjee, H.M., Wu, X.-H., Yu, Y.-T., Isobe, T., Takahashi, N., Min, J.(2016) Genes Dev 30: 2376-2390
- PubMed: 27881600
- DOI: https://doi.org/10.1101/gad.288340.116
- Primary Citation of Related Structures:
5GXH, 5GXI, 5TEE, 5TEF, 5THA - PubMed Abstract:
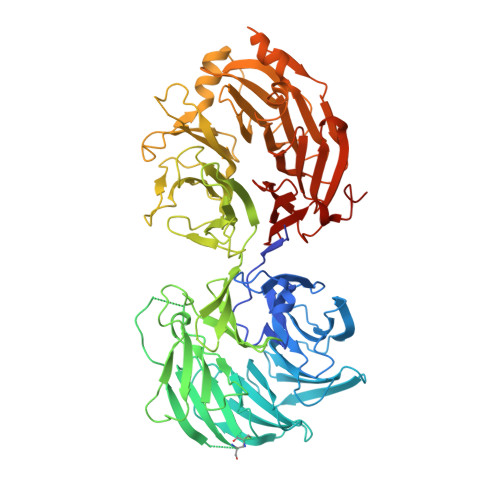
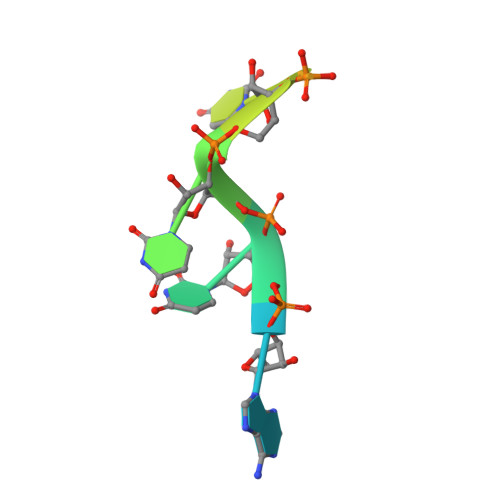
In cytoplasm, the survival of motor neuron (SMN) complex delivers pre-small nuclear RNAs (pre-snRNAs) to the heptameric Sm ring for the assembly of the ring complex on pre-snRNAs at the conserved Sm site [A(U) 4-6 G]. Gemin5, a WD40 protein component of the SMN complex, is responsible for recognizing pre-snRNAs. In addition, Gemin5 has been reported to specifically bind to the m 7 G cap. In this study, we show that the WD40 domain of Gemin5 is both necessary and sufficient for binding the Sm site of pre-snRNAs by isothermal titration calorimetry (ITC) and mutagenesis assays. We further determined the crystal structures of the WD40 domain of Gemin5 in complex with the Sm site or m 7 G cap of pre-snRNA, which reveal that the WD40 domain of Gemin5 recognizes the Sm site and m 7 G cap of pre-snRNAs via two distinct binding sites by respective base-specific interactions. In addition, we also uncovered a novel role of Gemin5 in escorting the truncated forms of U1 pre-snRNAs for proper disposal. Overall, the elucidated Gemin5 structures will contribute to a better understanding of Gemin5 in small nuclear ribonucleic protein (snRNP) biogenesis as well as, potentially, other cellular activities.
Organizational Affiliation:
Hefei National Laboratory for Physical Sciences at Microscale, Hefei Science Center of CAS, Chinese Academy of Science, School of Life Sciences, University of Science and Technology of China, Hefei, Anhui 230027, People's Republic of China.