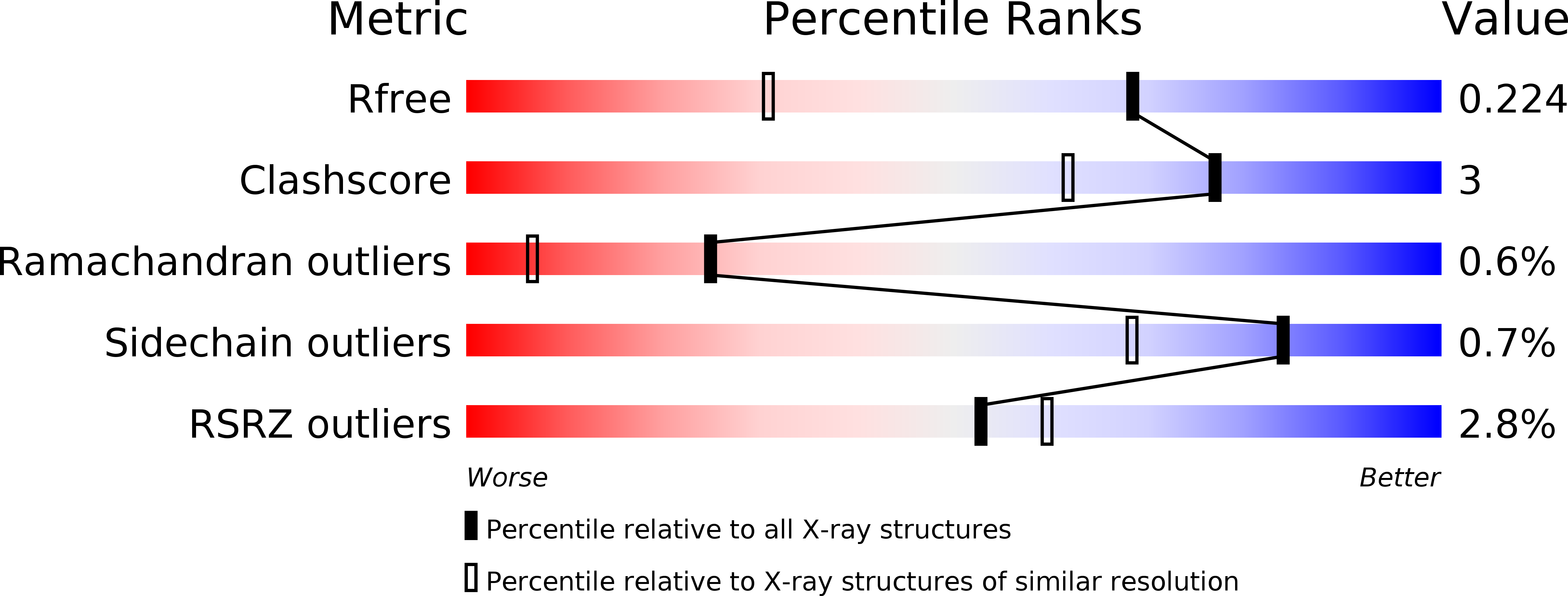
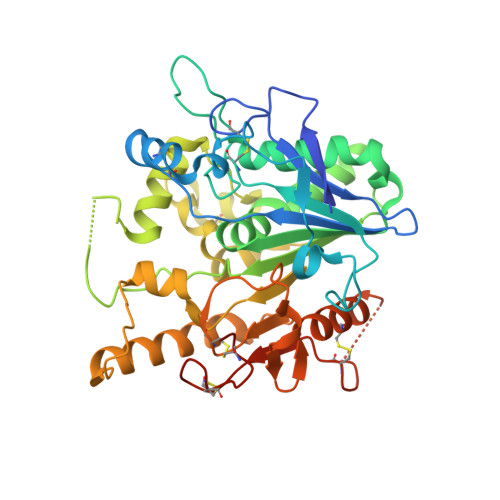
Caffeine inhibits Notum activity by binding at the catalytic pocket.
Zhao, Y., Ren, J., Hillier, J., Lu, W., Jones, E.Y.(2020) Commun Biol 3: 555-555
- PubMed: 33033363
- DOI: https://doi.org/10.1038/s42003-020-01286-5
- Primary Citation of Related Structures:
6TUZ, 6TV4 - PubMed Abstract:
Notum inhibits Wnt signalling via enzymatic delipidation of Wnt ligands. Restoration of Wnt signalling by small molecule inhibition of Notum may be of therapeutic benefit in a number of pathologies including Alzheimer's disease. Here we report Notum activity can be inhibited by caffeine (IC 50 19 µM), but not by demethylated caffeine metabolites: paraxanthine, theobromine and theophylline. Cellular luciferase assays show Notum-suppressed Wnt3a function can be restored by caffeine with an EC 50 of 46 µM. The dissociation constant (K d ) between Notum and caffeine is 85 µM as measured by surface plasmon resonance. High-resolution crystal structures of Notum complexes with caffeine and its minor metabolite theophylline show both compounds bind at the centre of the enzymatic pocket, overlapping the position of the natural substrate palmitoleic lipid, but using different binding modes. The structural information reported here may be of relevance for the design of more potent brain-accessible Notum inhibitors.
Organizational Affiliation:
Division of Structural Biology, Wellcome Centre for Human Genetics, University of Oxford, Oxford, OX3 7BN, UK. [email protected].