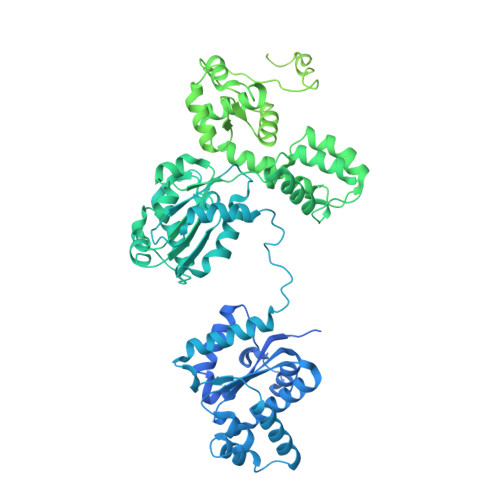
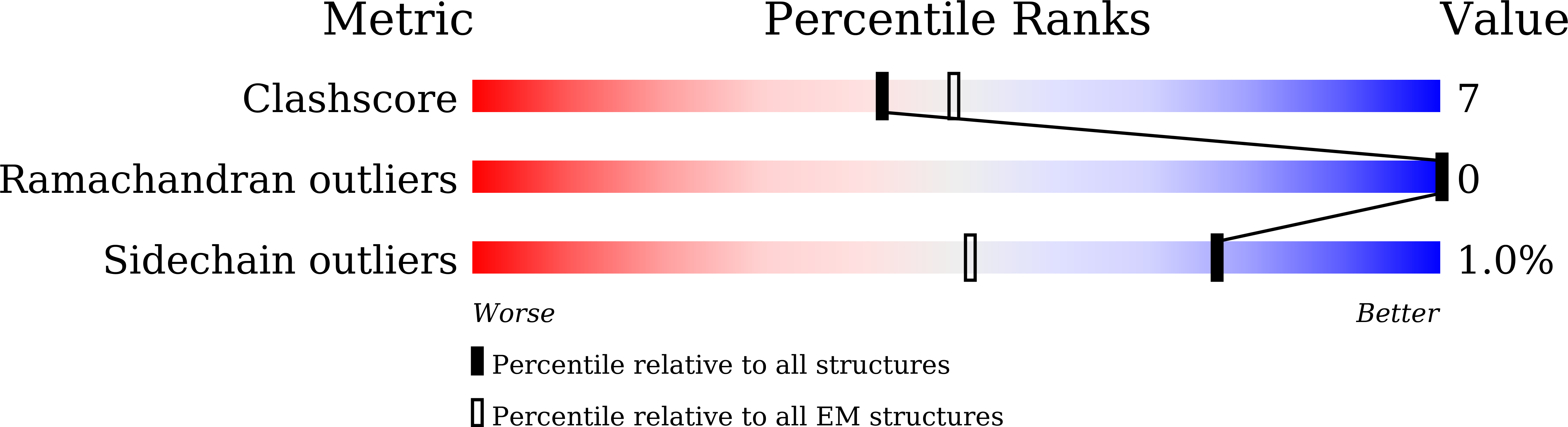Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme.
Ma, S., Lapin, D., Liu, L., Sun, Y., Song, W., Zhang, X., Logemann, E., Yu, D., Wang, J., Jirschitzka, J., Han, Z., Schulze-Lefert, P., Parker, J.E., Chai, J.(2020) Science 370
- PubMed: 33273071
- DOI: https://doi.org/10.1126/science.abe3069
- Primary Citation of Related Structures:
7CRB, 7CRC, 7DFV - PubMed Abstract:
Direct or indirect recognition of pathogen-derived effectors by plant nucleotide-binding leucine-rich repeat (LRR) receptors (NLRs) initiates innate immune responses. The Hyaloperonospora arabidopsidis effector ATR1 activates the N-terminal Toll-interleukin-1 receptor (TIR) domain of Arabidopsis NLR RPP1. We report a cryo-electron microscopy structure of RPP1 bound by ATR1. The structure reveals a C-terminal jelly roll/Ig-like domain (C-JID) for specific ATR1 recognition. Biochemical and functional analyses show that ATR1 binds to the C-JID and the LRRs to induce an RPP1 tetrameric assembly required for nicotinamide adenine dinucleotide hydrolase (NADase) activity. RPP1 tetramerization creates two potential active sites, each formed by an asymmetric TIR homodimer. Our data define the mechanism of direct effector recognition by a plant NLR leading to formation of a signaling-active holoenzyme.
Organizational Affiliation:
Beijing Advanced Innovation Center for Structural Biology, Tsinghua-Peking Center for Life Sciences, Centre for Plant Biology, School of Life Sciences, Tsinghua University, 100084 Beijing, China.