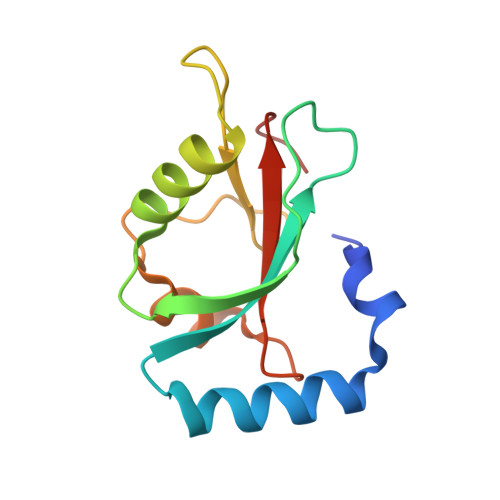
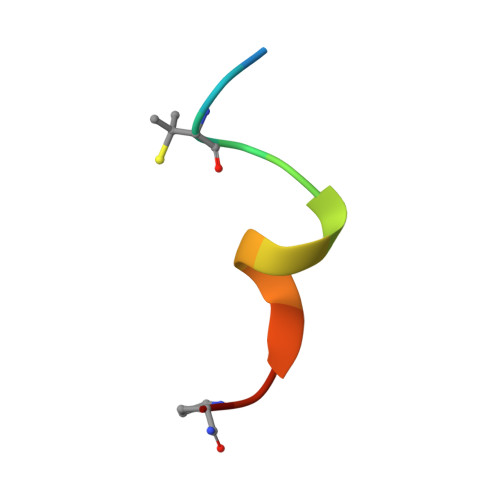Structure-Based Design of Stapled Peptides That Bind GABARAP and Inhibit Autophagy.
Brown, H., Chung, M., Uffing, A., Batistatou, N., Tsang, T., Doskocil, S., Mao, W., Willbold, D., Bast Jr., R.C., Lu, Z., Weiergraber, O.H., Kritzer, J.A.(2022) J Am Chem Soc 144: 14687-14697
- PubMed: 35917476
- DOI: https://doi.org/10.1021/jacs.2c04699
- Primary Citation of Related Structures:
7ZKR, 7ZL7 - PubMed Abstract:
The LC3/GABARAP family of proteins is involved in nearly every stage of autophagy. Inhibition of LC3/GABARAP proteins is a promising approach to blocking autophagy, which sensitizes advanced cancers to DNA-damaging chemotherapy. Here, we report the structure-based design of stapled peptides that inhibit GABARAP with nanomolar affinities. Small changes in staple structure produced stapled peptides with very different binding modes and functional differences in LC3/GABARAP paralog selectivity, ranging from highly GABARAP-specific to broad inhibition of both subfamilies. The stapled peptides exhibited considerable cytosolic penetration and resistance to biological degradation. They also reduced autophagic flux in cultured ovarian cancer cells and sensitized ovarian cancer cells to cisplatin. These small, potent stapled peptides represent promising autophagy-modulating compounds that can be developed as novel cancer therapeutics and novel mediators of targeted protein degradation.
Organizational Affiliation:
Department of Chemistry, Tufts University, Medford, Massachusetts 02155, United States.