Structural insights into the chloroplast protein import in land plants.
Liang, K., Jin, Z., Zhan, X., Li, Y., Xu, Q., Xie, Y., Yang, Y., Wang, S., Wu, J., Yan, Z.(2024) Cell
- PubMed: 39197452
- DOI: https://doi.org/10.1016/j.cell.2024.08.003
- Primary Citation of Related Structures:
8XKU, 8XKV - PubMed Abstract:
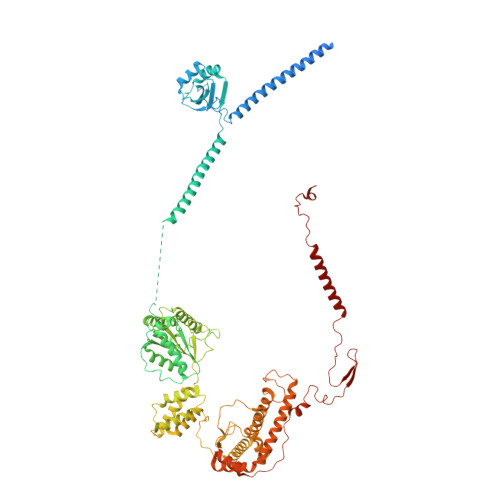
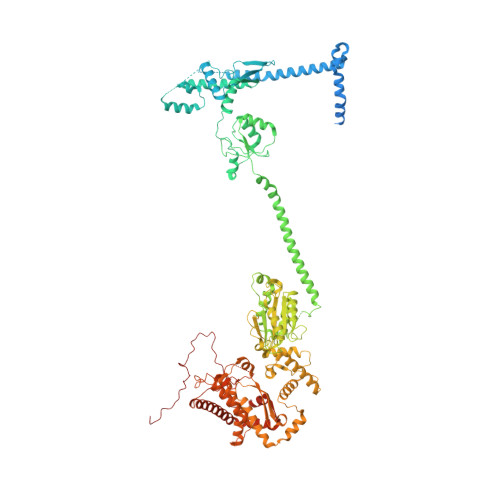
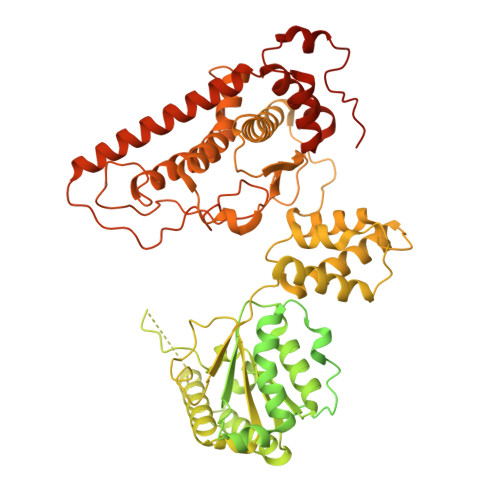
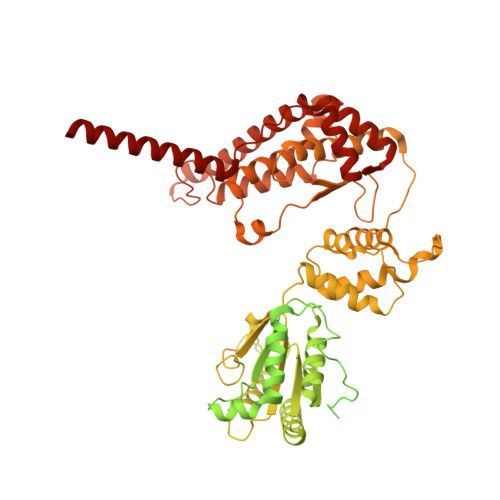
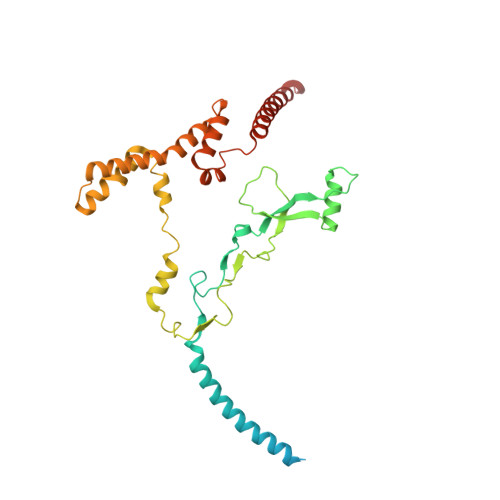
Chloroplast proteins are imported via the translocon at the outer chloroplast membrane (TOC)-translocon at the inner chloroplast membrane (TIC) supercomplex, driven by an ATPase motor. The Ycf2-FtsHi complex has been identified as the chloroplast import motor. However, its assembly and cooperation with the TIC complex during preprotein translocation remain unclear. Here, we present the structures of the Ycf2-FtsHi and TIC complexes from Arabidopsis and an ultracomplex formed between them from Pisum. The Ycf2-FtsHi structure reveals a heterohexameric AAA+ ATPase motor module with characteristic features. Four previously uncharacterized components of Ycf2-FtsHi were identified, which aid in complex assembly and anchoring of the motor module at a tilted angle relative to the membrane. When considering the structures of the TIC complex and the TIC-Ycf2-FtsHi ultracomplex together, it becomes evident that the tilted motor module of Ycf2-FtsHi enables its close contact with the TIC complex, thereby facilitating efficient preprotein translocation. Our study provides valuable structural insights into the chloroplast protein import process in land plants.
Organizational Affiliation:
Key Laboratory of Structural Biology of Zhejiang Province, School of Life Sciences, Westlake University, Hangzhou, Zhejiang 310024, China; Westlake Laboratory of Life Sciences and Biomedicine, Hangzhou, Zhejiang 310024, China; Institute of Biology, Westlake Institute for Advanced Study, Hangzhou, Zhejiang 310024, China.