News
Introducing Mol*
11/19 
Access Mol*, a new 3D molecular viewer developed in collaboration between RCSB PDB, PDBe, and CETIEC, from RCSB PDB and PDBe pages. This new viewer allows fast visualization of molecular structures and their corresponding data, along with high-quality rendering within the browser window.
The speed of Mol*, enabling visualization of huge structures in the browser and even on mobile devices, is achieved thanks to the use of binary CIF files, available as static files or delivered from the ModelServer and VolumeServer. This compressed format, delivering only the data that is required, ensures incredibly fast loading of both model and map data from PDB and EMDB entries. In addition to its speed, Mol* has a powerful rendering engine, enabling high quality visualization of molecular structures in various representations.
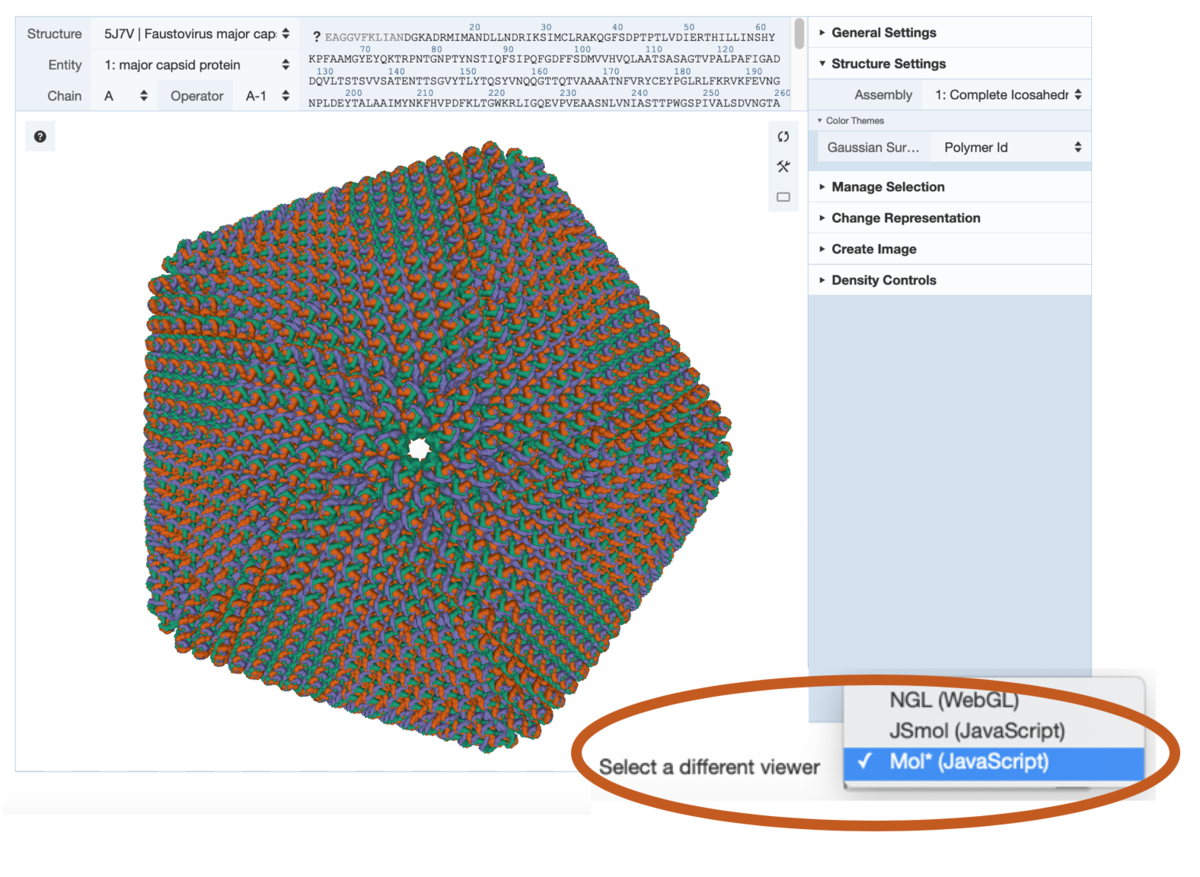 View for Faustovirus (PDB 5j7v) at RCSB.org. Toggle between 3D viewers at the bottom of the display.
View for Faustovirus (PDB 5j7v) at RCSB.org. Toggle between 3D viewers at the bottom of the display.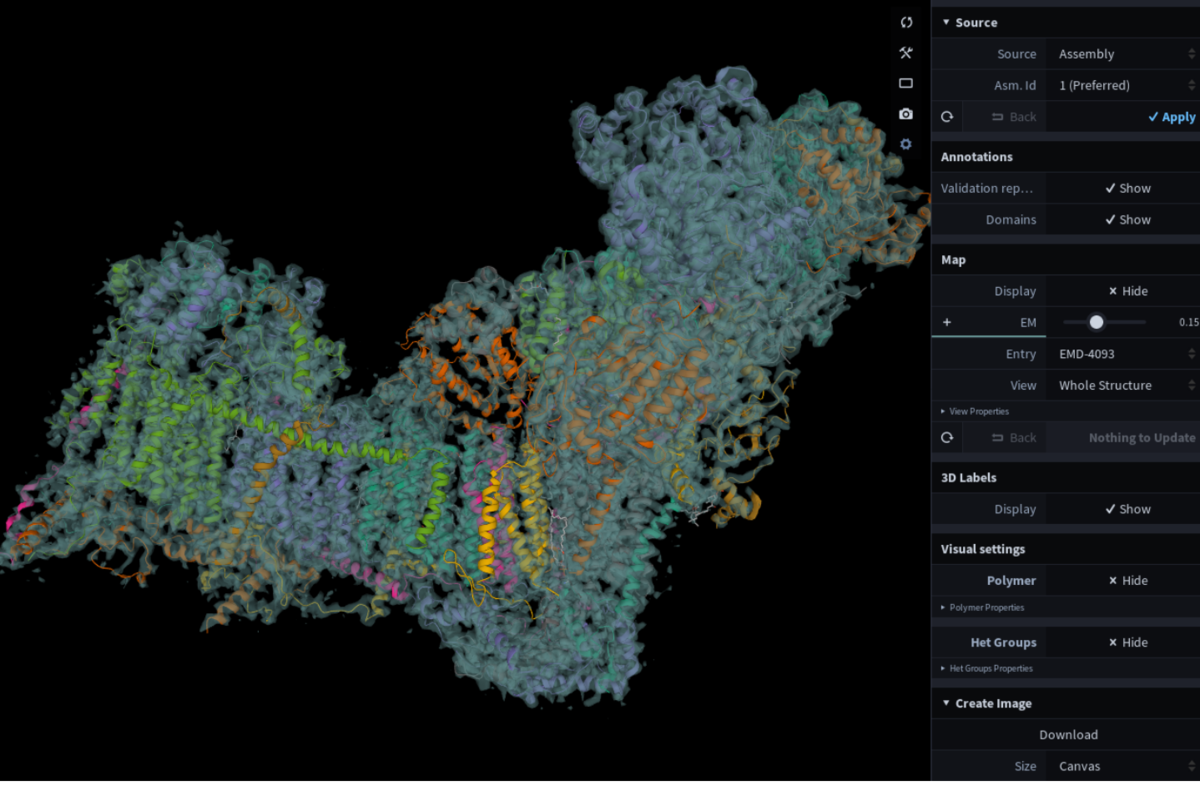 Mol* as displayed on the PDBe entry page for 5lnk. The associated EMDB map (EMD-4093) is automatically loaded upon opening the viewer.
Mol* as displayed on the PDBe entry page for 5lnk. The associated EMDB map (EMD-4093) is automatically loaded upon opening the viewer.
On the PDBe pages, Mol* has now replaced the LiteMol viewer in all instances, including on the PDBe search and entry pages (e.g. pdbe.org/5lnk/3d). Access Mol* from any 3D View tab for Structure Summary pages at RCSB.org. Both the LiteMol and NGL viewers at PDBe and RCSB PDB, respectively, will no longer be actively developed.
Mol* itself is an open-source project that provides a technology stack for data delivery and analysis tools of macromolecules. The source code is available on Github. Individual components (including the Viewer) of Mol* can be readily used in 3rd party applications. Additionally, the viewer is available as an easily embeddable web component through the PDBe PDB component library.
If you have any queries about the Mol* viewer implementation then either Contact RCSB PDB or press the ‘Feedback’ button at PDBe.












