News
View Validation in 3D
10/11
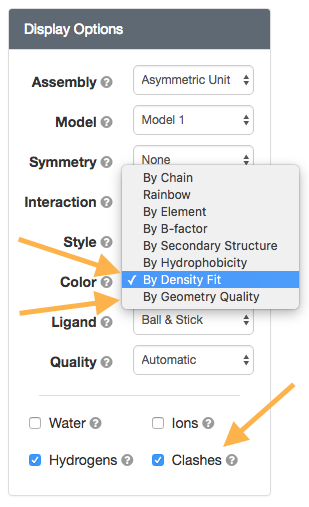 New NGL Viewer options
New NGL Viewer optionswwPDB Validation Reports are available for every entry to provide an assessment of the quality of a structure and highlight specific concerns by considering the model coordinates, experimental data, and fit between the two. RCSB PDB Structure Summary pages contain the “slider” graphic that provides a visual summary, and link to the full report (PDF).
New options in the NGL viewer map wwPDB Validation Report information onto the 3D structure. Clashes can be displayed as pink disks, and the full structure can be shown using “Geometry Quality” and “Density Fit” coloring schemes.
Color By Density Fit
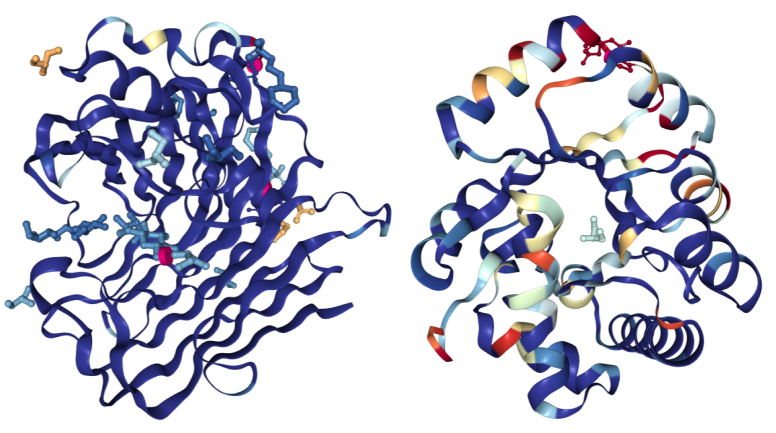
For structures determined using X-ray crystallography, the “Density Fit” scheme colors a structure according to the quality of agreement between the model and the experimental electron density. Blue indicates a good fit for a residue and red a bad fit. Residue coloring is determined using normalized Real Space R (RSRZ) for polymer residues and real space correlation coefficient (RSCC) for ligands. Colors range from red (RSRZ = -2 or RSCC = 0.678) - through white - to blue (RSRZ = 0 or RSCC = 1.0). The "Density Fit" color scheme is only available for models with deposited structure factors.
On the left is a structure of Endoglucanase A (PDB ID 3WY6) with a generally good fit; on the right a structure of Ribonuclease P protein component 3 (PDB ID 3WYZ), which has areas of more problematic fit. For details regarding RSRZ and RSCC, please consult the Validation Report User Guide for X-ray structures.
Color By Geometry Quality
The “Geometry Quality” coloring scheme colors each polymer residue and ligand molecule according to the number of geometric issues (blue for 0, yellow for 1, orange for 2, and red for 3 or more). Protein residues and nucleotides are colored per residue whereas ligand molecules are colored per atom. Possible geometric issues include steric clashes, Ramachandran or RNA backbone outliers, and sidechain conformation outliers.
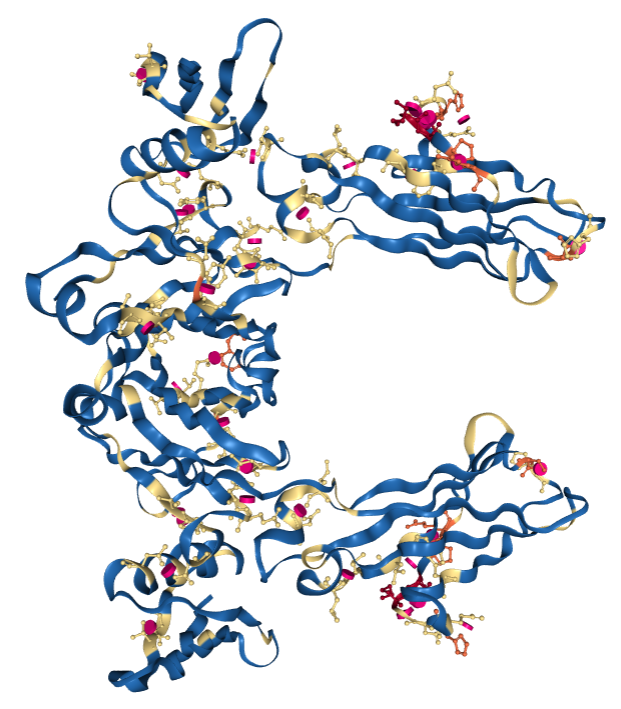 PDB entry 1FCC, a 3.2Å resolution structure with worse overall quality relative to all X-ray structures colored by Geometry Quality.
PDB entry 1FCC, a 3.2Å resolution structure with worse overall quality relative to all X-ray structures colored by Geometry Quality. Key: Color by Geometry Quality
Key: Color by Geometry QualityDisplay Clashes Between Atoms
With this option, clashes between pairs of atoms are displayed as pink discs, with the size of each disc reflecting the degree of van der Waals (vdW) overlap between the two atoms. Clash display is currently not available for structures comprising more than 10,000 residues.
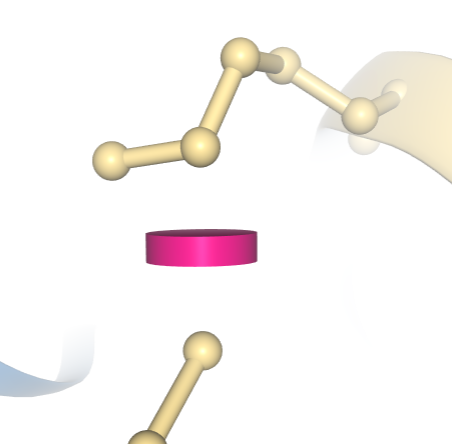 A clash between two atoms in PDB ID 1D66 is indicated by a pink disc, showing how much the atoms’ vdW spheres overlap. In this example, the structure has been colored using the “Geometry Quality” scheme, which indicates that this clash is the only geometric issue for these two residues.
A clash between two atoms in PDB ID 1D66 is indicated by a pink disc, showing how much the atoms’ vdW spheres overlap. In this example, the structure has been colored using the “Geometry Quality” scheme, which indicates that this clash is the only geometric issue for these two residues. Information on how these geometry quality criteria are calculated is available at wwPDB.org.







 Key: Color by Density Fit
Key: Color by Density Fit




