News
Explore Structural Biology with CSMs
10/09 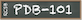
Ever since the first structures of proteins were determined, scientists have been searching for ways to predict the folding pattern of protein chains. After many years of study, several approaches have been successful. Homology modeling starts with a protein of known 3D structure and predicts the 3D structure of similar proteins based on the 1D sequence alignment. Newer methods, like AlphaFold2 and RosettaFold2, expand on this approach, using artificial intelligence/machine learning (AI/ML) to predict the structure based on a large database of known structures. Physics-based methods start from first principles and simulate the folding of proteins. Currently, homology modeling is highly effective for many well-folded proteins, AI/ML-based methods expand this to predict structures across entire proteomes, and physics-based methods are effective mostly for small proteins.
RCSB.org currently hosts a collection of more than one million computed structure models (CSMs) coming from the AlphaFold Database and the Model Archive. These data are delivered alongside more than 220,000 experimentally-determined PDB structures. Searching for both PDB structures and CSMs at RCSB.org can be turned on using the toggle located at the upper right corner of each RCSB.org web page.
 The experimental structure of chloroplast ATP synthase from spinach is shown on the left (PDB ID 6fkf). No experimental structure is currently available for the model organism Arabidopsis thaliana, but computed structure models of the individual protein subunits have been predicted using AlphaFold2, as shown on the right.
The experimental structure of chloroplast ATP synthase from spinach is shown on the left (PDB ID 6fkf). No experimental structure is currently available for the model organism Arabidopsis thaliana, but computed structure models of the individual protein subunits have been predicted using AlphaFold2, as shown on the right.











