News
COVID-19 Coronavirus Resources
02/06 
PDB data and related resources provide a starting point for structure-guided drug discovery and understanding of COVID-19.
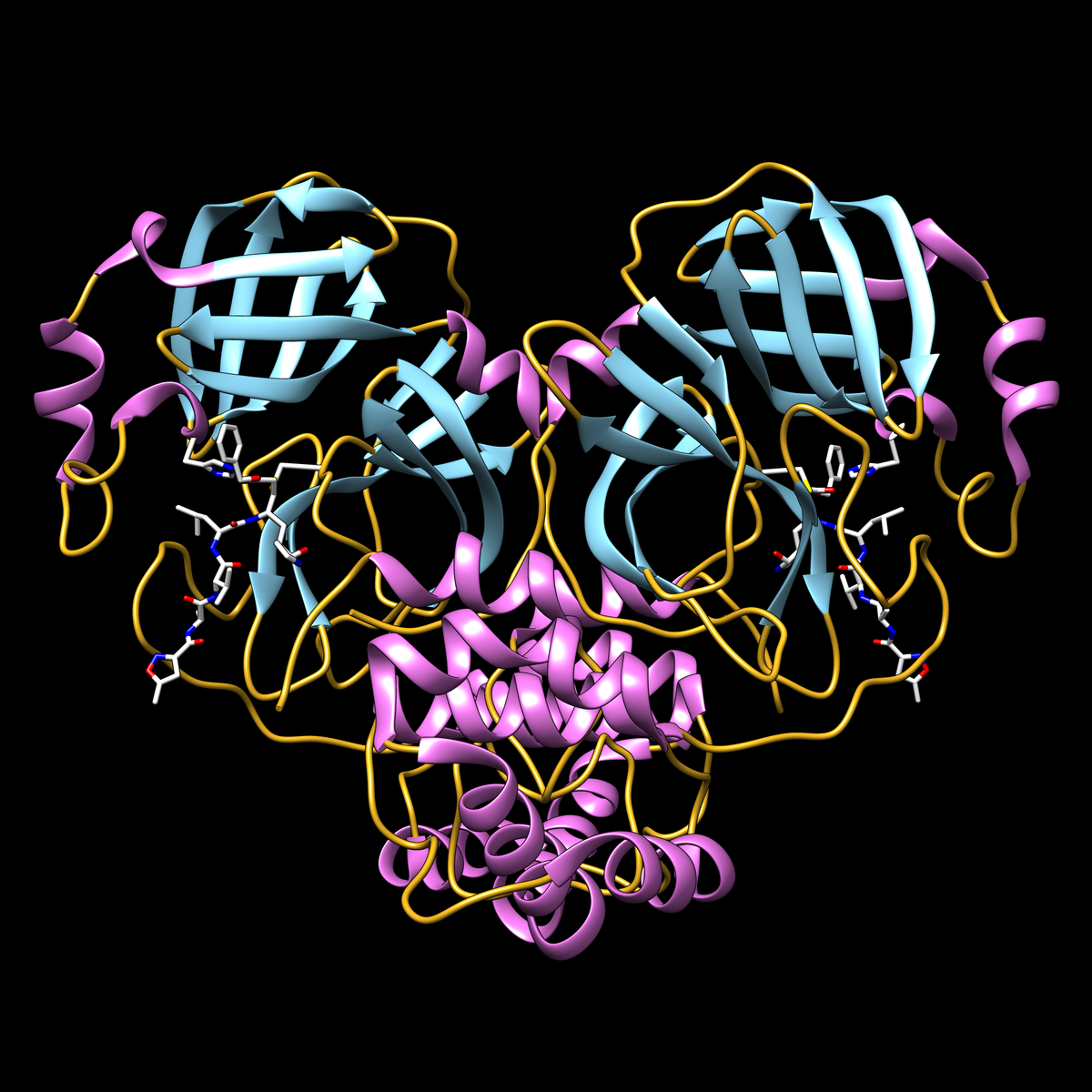
- PDB structure 6lu7: first high-resolution crystal structure of COVID-19 coronavirus 3CL hydrolase (Mpro) determined by Zihe Rao and Haitao Yang's research team at ShanghaiTech University (released February 5, 2020). doi: 10.2210/pdb6lu7/pdb
- wwPDB overview, including summary data for closely-related PDB structures (96% sequence identity; CSV) and other closely related SARS proteins, including a second viral protease, the RNA polymerase, the viral spike protein, a viral RNA, and other proteins (CSV)
- Molecule of the Month feature on Coronavirus Proteases
- Coronavirus painting
Images
Coronavirus, 2020. Illustration by David S. Goodsell, RCSB Protein Data Bank doi: 10.2210/rcsb_pdb/goodsell-gallery-019
Download high quality TIFF image
COVID-19 coronavirus main protease, with inhibitor in turquoise. Image from the February 2020 Molecule of the Month article.
Download high quality TIFF image
 Video: Fighting Coronavirus with Soap and related images
Video: Fighting Coronavirus with Soap and related imagesPDB Structures: Access all 29 COVID19-related PDB structures
 PDB structure 6lu7 Released 2020-02-05
PDB structure 6lu7 Released 2020-02-05Zhenming Jin, Xiaoyu Du, Yechun Xu, Yongqiang Deng, Meiqin Liu, Yao Zhao, Bing Zhang, Xiaofeng Li, Leike Zhang, Chao Peng, Yinkai Duan, Jing Yu, Lin Wang, Kailin Yang, Fengjiang Liu, Rendi Jiang, Xinglou Yang, Tian You, Xiaoce Liu, Xiuna Yang, Fang Bai, Hong Liu, Xiang Liu, Luke W. Guddat, Wenqing Xu, Gengfu Xiao, Chengfeng Qin, Zhengli Shi, Hualiang Jiang, Zihe Rao, Haitao Yang
Structure of Mpro from COVID-19 virus and discovery of its inhibitors. bioRxiv
doi: 10.1101/2020.02.26.964882
PDB structure 6vsb doi: 10.2210/pdb6vsb/pdb D. Wrapp, N. Wang, K.S. Corbett, J.A. Goldsmith, C.-L. Hsieh, O. Abiona, B.S. Graham, J.S. McLellan (2020) Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation Science doi: 10.1126/science.abb2507
Released 2020-02-26
Shown: Cryoelectron microscopy structure of 2019-nCoV (COVID-19) spike glycoprotein (PDB ID 6vsb) in the prefusion conformation, with the three subunits of the trimer in red, green, and blue, and glycosylation in yellow.
PDB structure 6lxt
Y. Zhu, F. Sun Structure of post fusion core of 2019-nCoV S2 subunit doi: 10.2210/pdb6lxt/pdb
Released 2020-02-26
PDB structure 6lvn
Y. Zhu, F. Sun Structure of the 2019-nCoV HR2 Domain doi: 10.2210/pdb6lvn/pdb
Released 2020-02-26
PDB structure 6vw1
J. Shang, G. Ye, K. Shi, Y.S. Wan, H. Aihara, F. Li Structural basis for receptor recognition by the novel coronavirus from Wuhan doi: 10.2210/pdb6vw1/pdb
Released 2020-03-04
PDB structure 6vww
Y. Kim, R. Jedrzejczak, N. Maltseva, M. Endres, A. Godzik, K. Michalska, A. Joachimiak, Center for Structural Genomics of Infectious Diseases Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 doi: 10.2210/pdb6vww/pdb
Released 2020-03-04
PDB structure 6y2e
L. Zhang, X. Sun, R. Hilgenfeld Crystal structure of the free enzyme of the SARS-CoV-2 (2019-nCoV) main protease doi: 10.2210/pdb6y2e/pdb
Released 2020-03-04
PDB structure 6y2f
L. Zhang, X. Sun, R. Hilgenfeld Crystal structure (monoclinic form) of the complex resulting from the reaction between SARS-CoV-2 (2019-nCoV) main protease and tert-butyl (1-((S)-1-(((S)-4-(benzylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)butan-2-yl)amino)-3-cyclopropyl-1-oxopropan-2-yl)-2-oxo-1,2-dihydropyridin-3-yl)carbamate (alpha-ketoamide 13b) doi: 10.2210/pdb6y2f/pdb
Released 2020-03-04
PDB structure 6y2g
L. Zhang, X. Sun, R. Hilgenfeld Crystal structure (orthorhombic form) of the complex resulting from the reaction between SARS-CoV-2 (2019-nCoV) main protease and tert-butyl (1-((S)-1-(((S)-4-(benzylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)butan-2-yl)amino)-3-cyclopropyl-1-oxopropan-2-yl)-2-oxo-1,2-dihydropyridin-3-yl)carbamate (alpha-ketoamide 13b) doi: 10.2210/pdb6y2g/pdb
Released 2020-03-04
COVID-19 main protease with unliganded active site and PanDDA analysis Deposition Group: G_1002135 (8 structures)
PDB structure 6Y84
C.D. Owen, P. Lukacik, C.M. Strain-Damerell, A. Douangamath, A.J. Powell, D. Fearon, J. Brandao-Neto, A.D. Crawshaw, D. Aragao, M. Williams, R. Flaig, D. Hall, K. McAauley, D.I. Stuart, F. von Delft, M.A. Walsh
COVID-19 main protease with unliganded active site doi: 10.2210/pdb6y84/pdb
Released 2020-03-11
PDB structure 5R7Y
D. Fearon, A.J. Powell, A. Douangamath, C.D. Owen, C. Wild, T. Krojer, P. Lukacik, C.M. Strain-Damerell, M.A. Walsh, F. von Delft
Crystal Structure of COVID-19 main protease in complex with Z45617795 doi: 10.2210/pdb5r7y/pdb
Released 2020-03-11
PDB structure 5R7Z
D. Fearon, A.J. Powell, A. Douangamath, C.D. Owen, C. Wild, T. Krojer, P. Lukacik, C.M. Strain-Damerell, M.A. Walsh, F. von Delft
Crystal Structure of COVID-19 main protease in complex with Z1220452176 doi: 10.2210/pdb5r7z/pdb
Released 2020-03-11
PDB structure 5R80
D. Fearon, A.J. Powell, A. Douangamath, C.D. Owen, C. Wild, T. Krojer, P. Lukacik, C.M. Strain-Damerell, M.A. Walsh, F. von Delft
Crystal Structure of COVID-19 main protease in complex with Z18197050 doi: 10.2210/pdb5r80/pdb
Released 2020-03-11
PDB structure 5R81
D. Fearon, A.J. Powell, A. Douangamath, C.D. Owen, C. Wild, T. Krojer, P. Lukacik, C.M. Strain-Damerell, M.A. Walsh, F. von Delft
Crystal Structure of COVID-19 main protease in complex with Z1367324110 doi: 10.2210/pdb5r81/pdb
Released 2020-03-11
PDB structure 5R82
D. Fearon, A.J. Powell, A. Douangamath, C.D. Owen, C. Wild, T. Krojer, P. Lukacik, C.M. Strain-Damerell, M.A. Walsh, F. von Delft
Crystal Structure of COVID-19 main protease in complex with Z219104216 doi: 10.2210/pdb5r82/pdb
Released 2020-03-11
PDB structure 5R83
D. Fearon, A.J. Powell, A. Douangamath, C.D. Owen, C. Wild, T. Krojer, P. Lukacik, C.M. Strain-Damerell, M.A. Walsh, F. von Delft
Crystal Structure of COVID-19 main protease in complex with Z44592329 doi: 10.2210/pdb5r83/pdb
Released 2020-03-11
PDB structure 5R84
D. Fearon, A.J. Powell, A. Douangamath, C.D. Owen, C. Wild, T. Krojer, P. Lukacik, C.M. Strain-Damerell, M.A. Walsh, F. von Delft
Crystal Structure of COVID-19 main protease in complex with Z31792168 doi: 10.2210/pdb5r84/pdb
Released 2020-03-11
PDB structure 6vyb SARS-CoV-2 spike ectodomain structure (open state)
Alexandra C. Walls, Young-Jun Park, M. Alejandra Tortorici, Abigail Wall, Andrew T. McGuire, David Veesler (2020) Structure, Function, and Antigenicity of the SARSCoV-2 Spike Glycoprotein Cell 180: 1-12 doi: 10.1016/j.cell.2020.02.058
Released 2020-03-11
PDB structure 6vxx Structure of the SARS-CoV-2 spike glycoprotein (closed state)
Alexandra C. Walls, Young-Jun Park, M. Alejandra Tortorici, Abigail Wall, Andrew T. McGuire, David Veesler (2020) Structure, Function, and Antigenicity of the SARSCoV-2 Spike Glycoprotein Cell 180: 1-12 doi: 10.1016/j.cell.2020.02.058
Released 2020-03-11
PDB structure 6m17
Renhong Yan, Yuanyuan Zhang, Yaning Li, Lu Xia, Yingying Guo, Qiang Zhou (2020) Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2 Science doi: 10.1126/science.abb2762
Released 2020-03-11
SARS-CoV2 binds to the receptor protein ACE2 on the surface of cells that it infects. By studying the interaction of the SARS-CoV-2 spike protein (S) to this receptor, researchers hope to design new inhibitors to block infection. Several structures have recently been released that reveal this interaction. 6m17 is the structure of the receptor binding domain (RBD) of SARS-CoV-2 S protein with the ACE2-B^0 AT1 complex, suggesting simultaneous binding of two S protein trimers to an ACE2 dimer. 6m18 and 6m1d are ACE2-B^0 AT1 complex structures in closed and open forms without RBD binding. B^0 AT1 stabilizes full length ACE2 in the structural studies.
PDB structure 6vyo
C. Chang, K. Michalska, R. Jedrzejczak, N. Maltseva, M. Endres, A. Godzik, Y. Kim, A. Joachimiak Crystal structure of RNA binding domain of nucleocapsid phosphoprotein from SARS coronavirus 2 doi: 10.2210/pdb6vyo/pdb
Released 2020-03-11
PDB structure 6w01
Y. Kim, R. Jedrzejczak, N. Maltseva, M. Endres, A. Godzik, K. Michalska, A. Joachimiak The 1.9 A Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with a Citrate doi: 10.2210/pdb6w01/pdb
Released 2020-03-11
PDB structure 6w02
K. Michalska, Y. Kim, R. Jedrzejczak, N. Maltseva, M. Endres, A. Mececar, A. Joachimiak Crystal Structure of ADP ribose phosphatase of NSP3 from SARS CoV-2 in the complex with ADP ribose doi: 10.2210/pdb6w02/pdb
Released 2020-03-11
PDB structure 6m03
B.Zhang, Y. Zhao, Z. Jin, X. Liu, H. Yang, Z. Rao, The crystal structure of COVID-19 main protease in apo form doi: 10.2210/pdb6m03/pdb
Released 2020-03-11
Released 2020-03-18
- 6LZG: Q.H. Wang, H. Song, J.X. Qi, Structure of novel coronavirus spike receptor-binding domain complexed with its receptor ACE2 doi: 10.2210/pdb6lzg/pdb
- 6M0J: X. Wang, Crystal structure of 2019-nCoV spike receptor-binding domain bound with ACE2 doi: 10.2210/pdb6m0j/pdb
- 6M3M: S. Chen, S. Kang, Structural insights of SARS-CoV-2 nucleocapsid protein RNA binding domain reveal potential unique drug targeting sites doi: 10.2210/pdb6m3m/pdb
- 6W4B: K. Tan, Y. Kim, R. Jedrzejczak, N. Maltseva, M. Endres, K. Michalska, A. Joachimiak, CSGID, The crystal structure of Nsp9 replicase protein of COVID-19 doi: 10.2210/pdb6w4b/pdb
- 6W4H: G. Minasov, L. Shuvalova, M. Rosas-Lemus, O. Kiryukhina, G. Wiersum, A. Godzik, L. Jaroszewski, P.J. Stogios, T. Skarina, K.J.F. Satchell, 1.80 Angstrom Resolution Crystal Structure of NSP16 - NSP10 Complex from SARS-CoV-2 doi: 10.2210/pdb6w4h/pdb