News
Real Time Structural Search of the PDB
08/11
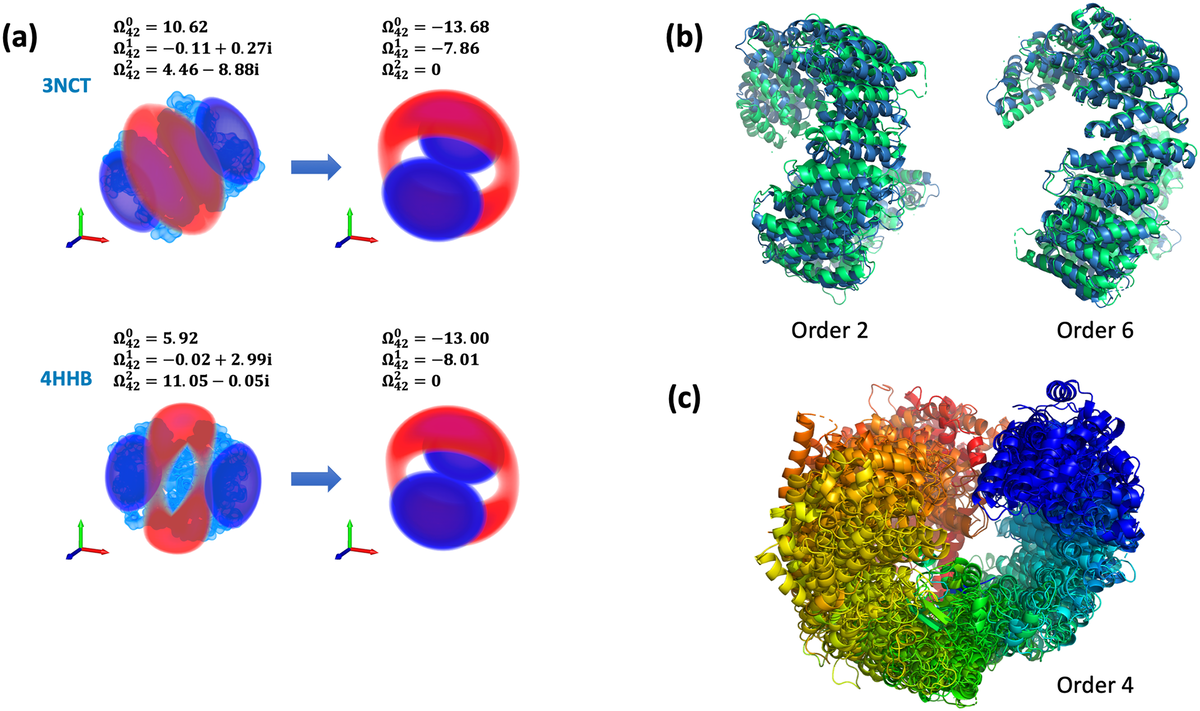
A new paper describes a novel method that detects similarity between protein shapes and that works equally fast for any size of proteins or assemblies.
Guzenko D, Burley SK, Duarte JM (2020) Real time structural search of the Protein Data Bank. PLoS Comput Biol 16(7): e1007970. doi: 10.1371/journal.pcbi.1007970
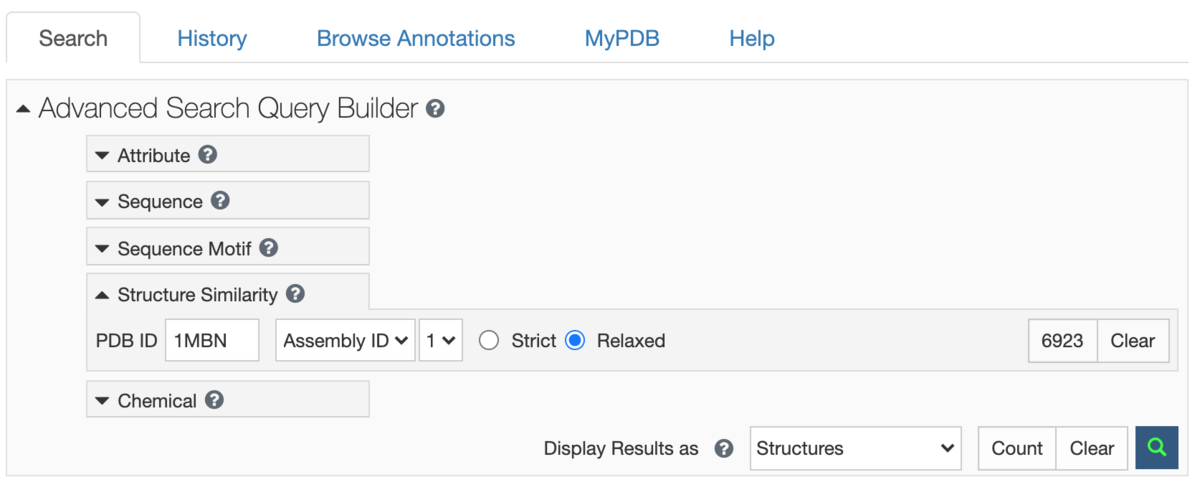
This method drives the Structure Similarity option in Advanced Search.
The system attempts to assess global 3D-shape similarity by using BioZernike descriptors that capture the global volumetric shape of the protein. Users can search for similar:
- polymeric chains to a given chain
- assemblies to a given assembly
Two modes of matching are available:
- Strict: returns matches that are all relevant but may not return more distant matches
- Relaxed: returns all similar matches, but may include some false positives